Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: defining a unique family of class C enzymes
- PMID: 15980372
- PMCID: PMC1168656
- DOI: 10.1128/AAC.49.7.2941-2948.2005
Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: defining a unique family of class C enzymes
Abstract
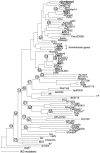
Acinetobacter spp. are emerging as opportunistic hospital pathogens that demonstrate resistance to many classes of antibiotics. In a metropolitan hospital in Cleveland, a clinical isolate of Acinetobacter baumannii that tested resistant to cefepime and ceftazidime (MIC = 32 microg/ml) was identified. Herein, we sought to determine the molecular basis for the extended-spectrum-cephalosporin resistance. Using analytical isoelectric focusing, a beta-lactamase with a pI of > or = 9.2 was detected. PCR amplification with specific A. baumannii cephalosporinase primers yielded a 1,152-bp product which, when sequenced, identified a novel 383-amino-acid class C enzyme. Expressed in Escherichia coli DH10B, this beta-lactamase demonstrated greater resistance against ceftazidime and cefotaxime than cefepime (4.0 microg/ml versus 0.06 microg/ml). The kinetic characteristics of this beta-lactamase were similar to other cephalosporinases found in Acinetobacter spp. In addition, this cephalosporinase was inhibited by meropenem, imipenem, ertapenem, and sulopenem (K(i) < 40 microM). The amino acid compositions of this novel enzyme and other class C beta-lactamases thus far described for A. baumannii, Acinetobacter genomic species 3, and Oligella urethralis in Europe and South Africa suggest that this cephalosporinase defines a unique family of class C enzymes. We propose a uniform designation for this family of cephalosporinases (Acinetobacter-derived cephalosporinases [ADC]) found in Acinetobacter spp. and identify this enzyme as ADC-7 beta-lactamase. The coalescence of Acinetobacter ampC beta-lactamases into a single common ancestor and the substantial phylogenetic distance separating them from other ampC genes support the logical value of developing a system of nomenclature for these Acinetobacter cephalosporinase genes.
Figures

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bonomo, R. A., J. Liu, Y. Chen, L. Ng, A. M. Hujer, and V. E. Anderson. 2001. Inactivation of CMY-2 β-lactamase by tazobactam: initial mass spectroscopic characterization. Biochim. Biophys. Acta 1547:196-205. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

