GlyProt: in silico glycosylation of proteins
- PMID: 15980456
- PMCID: PMC1160146
- DOI: 10.1093/nar/gki385
GlyProt: in silico glycosylation of proteins
Abstract
GlyProt (http://www.glycosciences.de/glyprot/) is a web-based tool that enables meaningful N-glycan conformations to be attached to all the spatially accessible potential N-glycosylation sites of a known three-dimensional (3D) protein structure. The probabilities of physicochemical properties such as mass, accessible surface and radius of gyration are calculated. The purpose of this service is to provide rapid access to reliable 3D models of glycoproteins, which can subsequently be refined by using more elaborate simulations and validated by comparing the generated models with experimental data.
Figures

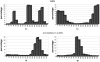
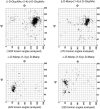


References
-
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. - PubMed
-
- Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta. 1999;1473:4–8. - PubMed
-
- Ben-Dor S., Esterman N., Rubin E. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14:95–101. - PubMed
-
- Luetteke T., Frank M., von der Lieth C.W. Data mining the protein data bank: automatic detection and assignment of carbohydrate structures. Carbohydr. Res. 2004;339:1015–1020. - PubMed
-
- Imberty A., Perez S. Structure, conformation, and dynamics of bioactive oligosaccharides: theoretical approaches and experimental validations. Chem. Rev. 2000;100:4567–4588. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

