ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures
- PMID: 15980475
- PMCID: PMC1160131
- DOI: 10.1093/nar/gki370
ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures
Abstract
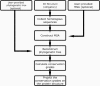
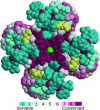
Key amino acid positions that are important for maintaining the 3D structure of a protein and/or its function(s), e.g. catalytic activity, binding to ligand, DNA or other proteins, are often under strong evolutionary constraints. Thus, the biological importance of a residue often correlates with its level of evolutionary conservation within the protein family. ConSurf (http://consurf.tau.ac.il/) is a web-based tool that automatically calculates evolutionary conservation scores and maps them on protein structures via a user-friendly interface. Structurally and functionally important regions in the protein typically appear as patches of evolutionarily conserved residues that are spatially close to each other. We present here version 3.0 of ConSurf. This new version includes an empirical Bayesian method for scoring conservation, which is more accurate than the maximum-likelihood method that was used in the earlier release. Various additional steps in the calculation can now be controlled by a number of advanced options, thus further improving the accuracy of the calculation. Moreover, ConSurf version 3.0 also includes a measure of confidence for the inferred amino acid conservation scores.
Figures
References
-
- Glaser F., Pupko T., Paz I., Bell R.E., Bechor-Shental D., Martz E., Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. - PubMed
-
- Mayrose I., Graur D., Ben-Tal N., Pupko T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol. Biol. Evol. 2004;21:1781–1791. - PubMed
-
- Pupko T., Bell R.E., Mayrose I., Glaser F., Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18:S71–S77. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

