Protein structure prediction servers at University College London
- PMID: 15980489
- PMCID: PMC1160171
- DOI: 10.1093/nar/gki410
Protein structure prediction servers at University College London
Abstract
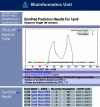
A number of state-of-the-art protein structure prediction servers have been developed by researchers working in the Bioinformatics Unit at University College London. The popular PSIPRED server allows users to perform secondary structure prediction, transmembrane topology prediction and protein fold recognition. More recent servers include DISOPRED for the prediction of protein dynamic disorder and DomPred for domain boundary prediction. These servers are available from our software home page at http://bioinf.cs.ucl.ac.uk/software.html.
Figures
References
-
- McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. - PubMed
-
- Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. - PubMed
-
- Rost B., Eyrich V.A. EVA: large-scale analysis of secondary structure prediction. Proteins. 2001;5:192–199. - PubMed
-
- Jones D.T. Do transmembrane protein superfolds exist? FEBS Lett. 1998;423:281–285. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources