BASys: a web server for automated bacterial genome annotation
- PMID: 15980511
- PMCID: PMC1160269
- DOI: 10.1093/nar/gki593
BASys: a web server for automated bacterial genome annotation
Abstract
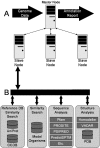
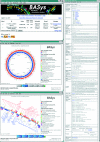
BASys (Bacterial Annotation System) is a web server that supports automated, in-depth annotation of bacterial genomic (chromosomal and plasmid) sequences. It accepts raw DNA sequence data and an optional list of gene identification information and provides extensive textual annotation and hyperlinked image output. BASys uses >30 programs to determine approximately 60 annotation subfields for each gene, including gene/protein name, GO function, COG function, possible paralogues and orthologues, molecular weight, isoelectric point, operon structure, subcellular localization, signal peptides, transmembrane regions, secondary structure, 3D structure, reactions and pathways. The depth and detail of a BASys annotation matches or exceeds that found in a standard SwissProt entry. BASys also generates colorful, clickable and fully zoomable maps of each query chromosome to permit rapid navigation and detailed visual analysis of all resulting gene annotations. The textual annotations and images that are provided by BASys can be generated in approximately 24 h for an average bacterial chromosome (5 Mb). BASys annotations may be viewed and downloaded anonymously or through a password protected access system. The BASys server and databases can also be downloaded and run locally. BASys is accessible at http://wishart.biology.ualberta.ca/basys.
Figures
References
-
- Andrade M.A., Brown N.P., Leroy C., Hoersch S., de Daruvar A., Reich C., Franchini A., Tamames J., Valencia A., Ouzounis C., Sander C. Automated genome sequence analysis and annotation. Bioinformatics. 1999;15:391–412. - PubMed
-
- Gaasterland T., Sensen C.W. MAGPIE: automated genome interpretation. Trends Genet. 1996;12:76–88. - PubMed
-
- Gordon P., Sensen C. Bluejay: a browser for linear units in Java. Proceedings of the 13th Annual International Symposium on High Performance Computing Systems and Applications; Kingston, ON, Canada. 1999. pp. 183–194.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

