SNP Cutter: a comprehensive tool for SNP PCR-RFLP assay design
- PMID: 15980518
- PMCID: PMC1160119
- DOI: 10.1093/nar/gki358
SNP Cutter: a comprehensive tool for SNP PCR-RFLP assay design
Abstract
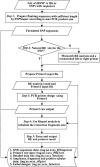
The Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) is a relatively simple and inexpensive method for genotyping single nucleotide polymorphisms (SNPs). It requires minimal investment in instrumentation. Here, we describe a web application, 'SNP Cutter,' which designs PCR-RFLP assays on a batch of SNPs from the human genome. NCBI dbSNP rs IDs or formatted SNPs are submitted into the SNP Cutter which then uses restriction enzymes from a pre-selected list to perform enzyme selection. The program is capable of designing primers for either natural PCR-RFLP or mismatch PCR-RFLP, depending on the SNP sequence data. SNP Cutter generates the information needed to evaluate and perform genotyping experiments, including a PCR primers list, sizes of original amplicons and different allelic fragment after enzyme digestion. Some output data is tab-delimited, therefore suitable for database archiving. The SNP Cut-ter is available at http://bioinfo.bsd.uchicago.edu/SNP_cutter.htm.
Figures

References
-
- Marnellos G. High-throughput SNP analysis for genetic association studies. Curr. Opin. Drug Discov. Devel. 2003;6:317–321. - PubMed
-
- Mooser V., Waterworth D.M., Isenhour T., Middleton L. Cardiovascular pharmacogenetics in the SNP era. J. Thromb. Haemost. 2003;1:1398–1402. - PubMed
-
- Hacia J.G., Fan J.B., Ryder O., Jin L., Edgemon K., Ghandour G., Mayer R.A., Sun B., Hsie L., Robbins C.M., et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nature Genet. 1999;22:164–167. - PubMed
-
- Syvanen A.C. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nature Rev. Genet. 2001;2:930–942. - PubMed
-
- Tsuchihashi Z., Dracopoli N.C. Progress in high throughput SNP genotyping methods. Pharmacogenomics. J. 2002;2:103–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

