Modeling Lactococcus lactis using a genome-scale flux model
- PMID: 15982422
- PMCID: PMC1185544
- DOI: 10.1186/1471-2180-5-39
Modeling Lactococcus lactis using a genome-scale flux model
Abstract
Background: Genome-scale flux models are useful tools to represent and analyze microbial metabolism. In this work we reconstructed the metabolic network of the lactic acid bacteria Lactococcus lactis and developed a genome-scale flux model able to simulate and analyze network capabilities and whole-cell function under aerobic and anaerobic continuous cultures. Flux balance analysis (FBA) and minimization of metabolic adjustment (MOMA) were used as modeling frameworks.
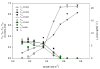
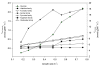
Results: The metabolic network was reconstructed using the annotated genome sequence from L. lactis ssp. lactis IL1403 together with physiological and biochemical information. The established network comprised a total of 621 reactions and 509 metabolites, representing the overall metabolism of L. lactis. Experimental data reported in the literature was used to fit the model to phenotypic observations. Regulatory constraints had to be included to simulate certain metabolic features, such as the shift from homo to heterolactic fermentation. A minimal medium for in silico growth was identified, indicating the requirement of four amino acids in addition to a sugar. Remarkably, de novo biosynthesis of four other amino acids was observed even when all amino acids were supplied, which is in good agreement with experimental observations. Additionally, enhanced metabolic engineering strategies for improved diacetyl producing strains were designed.
Conclusion: The L. lactis metabolic network can now be used for a better understanding of lactococcal metabolic capabilities and potential, for the design of enhanced metabolic engineering strategies and for integration with other types of 'omic' data, to assist in finding new information on cellular organization and function.
Figures


References
-
- van Niel EWJ, Hahn-Hagerdal B. Nutrient requirements of lactococci in defined growth media. Applied Microbiology and Biotechnology. 1999;52:617–627. doi: 10.1007/s002530051569. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

