Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels
- PMID: 15983066
- PMCID: PMC2212909
- DOI: 10.1084/jem.20050471
Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels
Abstract
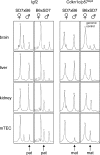
The role of central tolerance induction has recently been revised after the discovery of promiscuous expression of tissue-restricted self-antigens in the thymus. The extent of tissue representation afforded by this mechanism and its cellular and molecular regulation are barely defined. Here we show that medullary thymic epithelial cells (mTECs) are specialized to express a highly diverse set of genes representing essentially all tissues of the body. Most, but not all, of these genes are induced in functionally mature CD80(hi) mTECs. Although the autoimmune regulator (Aire) is responsible for inducing a large portion of this gene pool, numerous tissue-restricted genes are also up-regulated in mature mTECs in the absence of Aire. Promiscuously expressed genes tend to colocalize in clusters in the genome. Analysis of a particular gene locus revealed expression of clustered genes to be contiguous within such a cluster and to encompass both Aire-dependent and -independent genes. A role for epigenetic regulation is furthermore implied by the selective loss of imprinting of the insulin-like growth factor 2 gene in mTECs. Our data document a remarkable cellular and molecular specialization of the thymic stroma in order to mimic the transcriptome of multiple peripheral tissues and, thus, maximize the scope of central self-tolerance.
Figures






Comment in
-
Contrasting models of promiscuous gene expression by thymic epithelium.J Exp Med. 2005 Jul 4;202(1):15-9. doi: 10.1084/jem.20050976. Epub 2005 Jun 27. J Exp Med. 2005. PMID: 15983067 Free PMC article. Review.
References
-
- Kyewski, B., and J. Derbinski. 2004. Self-representation in the thymus: an extended view. Nat. Rev. Immunol. 4:688–698. - PubMed
-
- Sakaguchi, S. 2004. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 22:531–562. - PubMed
-
- Klein, L., M. Klugmann, K.A. Nave, V.K. Tuohy, and B. Kyewski. 2000. Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat. Med. 6:56–61. - PubMed
-
- Avichezer, D., R.S. Grajewski, C.C. Chan, M.J. Mattapallil, P.B. Silver, J.A. Raber, G.I. Liou, B. Wiggert, G.M. Lewis, L.A. Donoso, and R.R. Caspi. 2003. An immunologically privileged retinal antigen elicits tolerance: major role for central selection mechanisms. J. Exp. Med. 198:1665–1676. - PMC - PubMed
-
- Derbinski, J., A. Schulte, B. Kyewski, and L. Klein. 2001. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2:1032–1039. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

