Population fitness and the regulation of Escherichia coli genes by bacterial viruses
- PMID: 15984911
- PMCID: PMC1151598
- DOI: 10.1371/journal.pbio.0030229
Population fitness and the regulation of Escherichia coli genes by bacterial viruses
Abstract
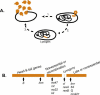
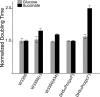
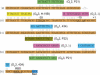
Temperate bacteriophage parasitize their host by integrating into the host genome where they provide additional genetic information that confers higher fitness on the host bacterium by protecting it against invasion by other bacteriophage, by increasing serum resistance, and by coding for toxins and adhesion factors that help the parasitized bacterium invade or evade its host. Here we ask if a temperate phage can also regulate host genes. We find several different host functions that are down-regulated in lysogens. The pckA gene, required for gluconeogenesis in all living systems, is regulated directly by the principal repressor of many different temperate prophage, the cI protein. cI binds to the regulatory region of pckA, thereby shutting down pckA transcription. The pckA regulatory region has target sequences for many other temperate phage repressors, and thus we suggest that down-regulation of the host pckA pathway increases lysogen fitness by lowering the growth rate of lysogens in energy-poor environments, perhaps as an adaptive response to the host predation system or as an aspect of lysogeny that must be offset by down-regulating pckA.
Figures


References
-
- Ackermann H-W, DuBow MS. Viruses of prokaryotes. Boca Raton (Florida): CRC Press; 1987. 480 pp.
-
- Hendrix RW. Lambda II. Cold Spring Harbor (New York): Cold Spring Harbor Laboratory Press; 1983. 694 pp.
-
- Adams MH. Bacteriophages. New York: Interscience Publishers; 1959. 592 pp.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

