Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs
- PMID: 15985150
- PMCID: PMC1175996
- DOI: 10.1186/jbiol24
Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs
Abstract
Background: During the translation of mRNA into polypeptide, elongation factor G (EF-G) catalyzes the translocation of peptidyl-tRNA from the A site to the P site of the ribosome. According to the 'classical' model, EF-G in the GTP-bound form promotes translocation, while hydrolysis of the bound GTP promotes dissociation of the factor from the post-translocation ribosome. According to a more recent model, EF-G operates like a 'motor protein' and drives translocation of the peptidyl-tRNA after GTP hydrolysis. In both the classical and motor protein models, GDP-to-GTP exchange is assumed to occur spontaneously on 'free' EF-G even in the absence of a guanine-nucleotide exchange factor (GEF).
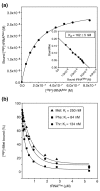
Results: We have made a number of findings that challenge both models. First, free EF-G in the cell is likely to be in the GDP-bound form. Second, the ribosome acts as the GEF for EF-G. Third, after guanine-nucleotide exchange, EF-G in the GTP-bound form moves the tRNA2-mRNA complex to an intermediate translocation state in which the mRNA is partially translocated. Fourth, subsequent accommodation of the tRNA2-mRNA complex in the post-translocation state requires GTP hydrolysis.
Conclusion: These results, in conjunction with previously published cryo-electron microscopy reconstructions of the ribosome in various functional states, suggest a novel mechanism for translocation of tRNAs on the ribosome by EF-G. Our observations suggest that the ribosome is a universal guanosine-nucleotide exchange factor for EF-G as previously shown for the class-II peptide-release factor 3.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

