An inactivated nuclease-like domain in RecC with novel function: implications for evolution
- PMID: 15985153
- PMCID: PMC1185551
- DOI: 10.1186/1472-6807-5-9
An inactivated nuclease-like domain in RecC with novel function: implications for evolution
Abstract
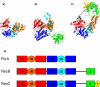
Background: The PD-(D/E)xK superfamily, containing a wide variety of other exo- and endonucleases, is a notable example of general function conservation in the face of extreme sequence and structural variation. Almost all members employ a small number of shared conserved residues to bind catalytically essential metal ions and thereby effect DNA cleavage. The crystal structure of the RecBCD prokaryotic DNA repair machinery shows that RecB contains such a nuclease domain at its C-terminus. The RecC C-terminal region was reported as having a novel fold.
Results: The RecC C-terminal region can be divided into an alpha/beta domain and a smaller alpha-helical bundle domain. Here we show that the alpha/beta domain is homologous to the RecB nuclease domain but lacks the features necessary for catalysis. Instead, the domain has a novel function within the nuclease superfamily--providing a hoop through which single-stranded DNA passes. Comparison with other structures of nuclease domains bound to DNA reveals strikingly different modes of ligand binding. The alpha-helical bundle domain contributes the pin which splits the DNA duplex.
Conclusion: The demonstrated homology of RecB and RecC shows how evolution acted to produce the present RecBCD complex through aggregation of new domains as well as functional divergence and structural redeployment of existing domains. Distantly homologous nuclease(-like) domains bind DNA in highly diverse manners.
Figures
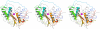
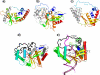

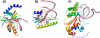
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

