Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants
- PMID: 15987780
- PMCID: PMC1174986
- DOI: 10.1073/pnas.0501957102
Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants
Abstract
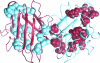
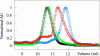
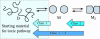
Amyotrophic lateral sclerosis is a neurodegenerative syndrome associated with 114 mutations in the gene encoding the cytosolic homodimeric enzyme Cu/Zn superoxide dismutase (SOD). In this article, we report that amyotrophic lateral sclerosis-associated SOD mutations with distinctly different disease progression can be rationalized in terms of their folding patterns. The mutations are found to perturb the protein in multiple ways; they destabilize the precursor monomers (class 1), weaken the dimer interface (class 2), or both at the same time (class 1 + 2). A shared feature of the mutational perturbations is a shift of the folding equilibrium toward poorly structured SOD monomers. We observed a link, coupled to the altered folding patterns, between protein stability, net charge, and survival time for the patients carrying the mutations.
Figures
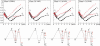

References
-
- Rosen, D. R., Siddique, T., Patterson, D., Figlewicz, D. A., Sapp, P., Hentati, A., Donaldson, D., Goto, J., O'Regan, J. P., Deng, H. X., et al. (1993) Nature 362, 59–62. - PubMed
-
- McCord, J. M. & Fridovich, I. (1969) J. Biol. Chem. 244, 6049–6055. - PubMed
-
- Brown, R. H., Jr., (1998) Nat. Med. 4, 1362–1364. - PubMed
-
- Shinder, G. A., Lacourse, M. C., Minotti, S. & Durham, H. D. (2001) J. Biol. Chem. 276, 12791–12796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

