An end-healing enzyme from Clostridium thermocellum with 5' kinase, 2',3' phosphatase, and adenylyltransferase activities
- PMID: 15987807
- PMCID: PMC1370810
- DOI: 10.1261/rna.2690505
An end-healing enzyme from Clostridium thermocellum with 5' kinase, 2',3' phosphatase, and adenylyltransferase activities
Abstract
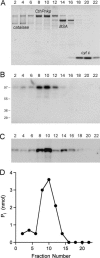
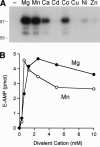
We identify and characterize an end-healing enzyme, CthPnkp, from Clostridium thermocellum that catalyzes the phosphorylation of 5'-OH termini of DNA or RNA polynucleotides and the dephosphorylation of 2',3' cyclic phosphate, 2'-phosphate, and 3'-phosphate ribonucleotides. CthPnkp also catalyzes an autoadenylylation reaction via a polynucleotide ligase-type mechanism. These characteristics are consistent with a role in end-healing during RNA or DNA repair. CthPnkp is a homodimer of an 870-amino-acid polypeptide composed of three catalytic domains: an N-terminal module that resembles the polynucleotide kinase domain of bacteriophage T4 Pnkp, a central metal-dependent phosphoesterase module, and a C-terminal module that resembles the nucleotidyl transferase domain of polynucleotide ligases. The distinctive feature of CthPnkp vis-à-vis known RNA repair enzymes is that its 3' end modification component belongs to the calcineurin-type phosphatase superfamily. It contains putative counterparts of the amino acids that form the dinuclear metal-binding site and the phosphate-binding site of bacteriophage lambda phosphatase. As with lambda phosphatase, the 2',3' cAMP phosphatase activity of CthPnkp is specifically dependent on nickel or manganese. We identify homologs of CthPnkp in other bacterial proteomes.
Figures





Similar articles
-
Mechanism of the phosphatase component of Clostridium thermocellum polynucleotide kinase-phosphatase.RNA. 2006 Jan;12(1):73-82. doi: 10.1261/rna.2196406. Epub 2005 Nov 21. RNA. 2006. PMID: 16301605 Free PMC article.
-
Characterization of the 2',3' cyclic phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase and bacteriophage lambda phosphatase.Nucleic Acids Res. 2007;35(22):7721-32. doi: 10.1093/nar/gkm868. Epub 2007 Nov 5. Nucleic Acids Res. 2007. PMID: 17986465 Free PMC article.
-
Reprogramming the tRNA-splicing activity of a bacterial RNA repair enzyme.Nucleic Acids Res. 2007;35(11):3624-30. doi: 10.1093/nar/gkm110. Epub 2007 May 8. Nucleic Acids Res. 2007. PMID: 17488852 Free PMC article.
-
Distinct enzymic functional groups are required for the phosphomonoesterase and phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase/phosphatase.J Biol Chem. 2006 Jul 14;281(28):19251-9. doi: 10.1074/jbc.M602549200. Epub 2006 May 4. J Biol Chem. 2006. PMID: 16675457
-
The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases.Curr Opin Struct Biol. 2004 Dec;14(6):757-64. doi: 10.1016/j.sbi.2004.10.006. Curr Opin Struct Biol. 2004. PMID: 15582400 Review.
Cited by
-
Reconstitution and structure of a bacterial Pnkp1-Rnl-Hen1 RNA repair complex.Nat Commun. 2015 Apr 17;6:6876. doi: 10.1038/ncomms7876. Nat Commun. 2015. PMID: 25882814 Free PMC article.
-
RtcB is the RNA ligase component of an Escherichia coli RNA repair operon.J Biol Chem. 2011 Mar 11;286(10):7727-7731. doi: 10.1074/jbc.C111.219022. Epub 2011 Jan 11. J Biol Chem. 2011. PMID: 21224389 Free PMC article.
-
A phosphate-binding histidine of binuclear metallophosphodiesterase enzymes is a determinant of 2',3'-cyclic nucleotide phosphodiesterase activity.J Biol Chem. 2008 Nov 7;283(45):30942-9. doi: 10.1074/jbc.M805064200. Epub 2008 Aug 28. J Biol Chem. 2008. PMID: 18757371 Free PMC article.
-
Active site mapping and substrate specificity of bacterial Hen1, a manganese-dependent 3' terminal RNA ribose 2'O-methyltransferase.RNA. 2011 Mar;17(3):429-38. doi: 10.1261/rna.2500711. Epub 2011 Jan 4. RNA. 2011. PMID: 21205839 Free PMC article.
-
SPICE: discovery of phenotype-determining component interplays.BMC Syst Biol. 2012 May 14;6:40. doi: 10.1186/1752-0509-6-40. BMC Syst Biol. 2012. PMID: 22583800 Free PMC article.
References
-
- Apostol, B.L., Westaway, S.K., Abelson, J., and Greer, C.L. 1991. Deletion analysis of a multifunctional yeast tRNA ligase polypeptide: Identification of essential and dispensable functional domains. J. Biol. Chem. 266: 7445–7455. - PubMed
-
- Aravind, L. and Koonin, E.V. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23: 469–472. - PubMed
-
- Bernstein, N.K., Williams, R.S., Rakovszky, M.L., Cui, D., Green, R., Karimi-Busheri, F., Mani, R.S., Galicia, S., Koch, C.A., Cass, C.E., et al. 2005. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell 17: 657–670. - PubMed
-
- Blondal, T., Hjorleifsdottir, S., Aevarsson, A., Fridjonsson, O.H., Skirnisdottir, S., Wheat, J.O., Hermannsdottir, A.G., Hreggvidsson, G.O., Smith, A.V., and Kristjansson, J.K. 2005. Characterization of a 5′-polynucleotide kinase/3′-phosphatase from bacteriophage RM378. J. Biol. Chem. 280: 5188–5194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
