A scalable, GFP-based pipeline for membrane protein overexpression screening and purification
- PMID: 15987891
- PMCID: PMC2279312
- DOI: 10.1110/ps.051466205
A scalable, GFP-based pipeline for membrane protein overexpression screening and purification
Abstract
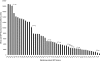
We describe a generic, GFP-based pipeline for membrane protein overexpression and purification in Escherichia coli. We exemplify the use of the pipeline by the identification and characterization of E. coli YedZ, a new, membrane-integral flavocytochrome. The approach is scalable and suitable for high-throughput applications. The GFP-based pipeline will facilitate the characterization of the E. coli membrane proteome and serves as an important reference for the characterization of other membrane proteomes.
Figures

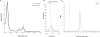
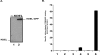
References
-
- Barber, M.J., Desai, S.K., Marohnic, C.C., Hernandez, H.H., and Pollock, V.V. 2002. Synthesis and bacterial expression of a gene encoding the heme domain of assimilatory nitrate reductase. Arch. Biochem. Biophys. 402 38–50. - PubMed
-
- Burch, H.B. 1957. Fluorometric assay of FAD, FMN, and riboflavin. Methods Enzymol. 3 960–962.
-
- Daley, D.O., Rapp, M., Granseth, E., Melén, K., Drew, D., and von Heijne, G. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308 1321–1323. - PubMed
-
- Drew, D.E., von Heijne, G., Nordlund, P., and de Gier, J.W. 2001. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 507 220–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

