Boundaries and physical characterization of a new domain shared between mammalian 53BP1 and yeast Rad9 checkpoint proteins
- PMID: 15987907
- PMCID: PMC2253359
- DOI: 10.1110/ps.041305205
Boundaries and physical characterization of a new domain shared between mammalian 53BP1 and yeast Rad9 checkpoint proteins
Abstract
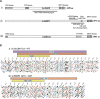
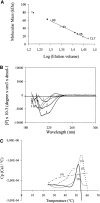
Eukaryotic cells have evolved DNA damage checkpoints in response to genome damage. They delay the cell cycle and activate repair mechanisms. The kinases at the heart of these pathways and the accessory proteins, which localize to DNA lesions and regulate kinase activation, are conserved from yeast to mammals. For Saccharomyces cerevisiae Rad9, a key adaptor protein in DNA damage checkpoint pathways, no clear human ortholog has yet been described in mammals. Rad9, however, shares localized homology with both human BRCA1 and 53BP1 since they all contain tandem C-terminal BRCT (BRCA1 C-terminal) motifs. 53BP1 is also a key mediator in DNA damage signaling required for cell cycle arrest, which has just been reported to possess a tandem Tudor repeat upstream of the BRCT motifs. Here we show that the major globular domain upstream of yeast Rad9 BRCT domains is structurally extremely similar to the Tudor domains recently resolved for 53BP1 and SMN. By expressing several fragments encompassing the Tudor-related motif and characterizing them using various physical methods, we isolated the independently folded unit for yeast Rad9. As in 53BP1, the domain corresponds to the SMN Tudor motif plus the contiguous HCA predicted structure region at the C terminus. These domains may help to further elucidate the structural and functional features of these two proteins and improve knowledge of the proteins involved in DNA damage.
Figures

Similar articles
-
The checkpoint Saccharomyces cerevisiae Rad9 protein contains a tandem tudor domain that recognizes DNA.Nucleic Acids Res. 2007;35(17):5898-912. doi: 10.1093/nar/gkm607. Epub 2007 Aug 28. Nucleic Acids Res. 2007. PMID: 17726056 Free PMC article.
-
A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins.FASEB J. 1997 Jan;11(1):68-76. FASEB J. 1997. PMID: 9034168
-
The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding.Structure. 2004 Sep;12(9):1551-62. doi: 10.1016/j.str.2004.06.014. Structure. 2004. PMID: 15341721
-
Interactions between BRCT repeats and phosphoproteins: tangled up in two.Trends Biochem Sci. 2004 Nov;29(11):579-85. doi: 10.1016/j.tibs.2004.09.010. Trends Biochem Sci. 2004. PMID: 15501676 Review.
-
ATM signaling and 53BP1.Radiother Oncol. 2005 Aug;76(2):119-22. doi: 10.1016/j.radonc.2005.06.026. Radiother Oncol. 2005. PMID: 16024119 Review.
Cited by
-
The checkpoint Saccharomyces cerevisiae Rad9 protein contains a tandem tudor domain that recognizes DNA.Nucleic Acids Res. 2007;35(17):5898-912. doi: 10.1093/nar/gkm607. Epub 2007 Aug 28. Nucleic Acids Res. 2007. PMID: 17726056 Free PMC article.
-
The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18.Genetics. 2006 Aug;173(4):1951-68. doi: 10.1534/genetics.106.057794. Epub 2006 Jun 18. Genetics. 2006. PMID: 16783014 Free PMC article.
-
An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks.Mol Cell Biol. 2009 Feb;29(4):1050-8. doi: 10.1128/MCB.01011-08. Epub 2008 Dec 8. Mol Cell Biol. 2009. PMID: 19064641 Free PMC article.
-
Maintenance of the DNA-damage checkpoint requires DNA-damage-induced mediator protein oligomerization.Mol Cell. 2009 Jan 30;33(2):147-59. doi: 10.1016/j.molcel.2008.12.022. Mol Cell. 2009. PMID: 19187758 Free PMC article.
References
-
- Baneyx, F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10 411–421. - PubMed
-
- Bork, P., Hofmann, K., Bucher, P., Neuwald, A.F., Altschul, S.F., and Koonin, E.V. 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11 68–76. - PubMed
-
- Callebaut, I. and Mornon, J.P. 1997a. From BRCA1 to RAP1: A widespread BRCT module closely associated with DNA repair. FEBS Lett. 400 25–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

