Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-kappaB and AP-1
- PMID: 15988009
- PMCID: PMC1168815
- DOI: 10.1128/MCB.25.14.5933-5946.2005
Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-kappaB and AP-1
Abstract
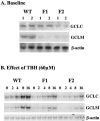
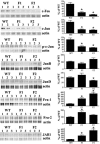
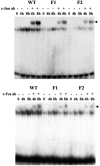
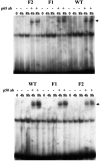
Glutamate-cysteine ligase catalytic subunit (GCLC) is regulated transcriptionally by Nrf1 and Nrf2. tert-Butylhydroquinone (TBH) induces human GCLC via Nrf2-mediated trans activation of the antioxidant-responsive element (ARE). Interestingly, TBH also induces rat GCLC, but the rat GCLC promoter lacks ARE. This study examined the role of Nrf1 and Nrf2 in the transcriptional regulation of rat GCLC. The baseline and TBH-mediated increase in GCLC mRNA levels and rat GCLC promoter activity were lower in Nrf1 and Nrf2 null (F1 and F2) fibroblasts than in wild-type cells. The basal protein and mRNA levels and nuclear binding activities of c-Jun, c-Fos, p50, and p65 were lower in F1 and F2 cells and exhibited a blunted response to TBH. Lower c-Jun and p65 expression also occurs in Nrf2 null livers. Levels of other AP-1 and NF-kappaB family members were either unaffected (i.e., JunB) or increased (i.e., Fra-1). Overexpression of Nrf1 and Nrf2 in respective cells restored the rat GCLC promoter activity and response to TBH but not if the AP-1 and NF-kappaB binding sites were mutated. Fra-1 overexpression lowered endogenous GCLC expression and rat GCLC promoter activity, while Fra-1 antisense had the opposite effects. In conclusion, Nrf1 and Nrf2 regulate rat GCLC promoter by modulating the expression of key AP-1 and NF-kappaB family members.
Figures








Similar articles
-
Tumour necrosis factor alpha induces co-ordinated activation of rat GSH synthetic enzymes via nuclear factor kappaB and activator protein-1.Biochem J. 2005 Oct 15;391(Pt 2):399-408. doi: 10.1042/BJ20050795. Biochem J. 2005. PMID: 16011481 Free PMC article.
-
Identification and characterization of an Nrf2-mediated ARE upstream of the rat glutamate cysteine ligase catalytic subunit gene (GCLC).J Cell Biochem. 2009 Aug 1;107(5):944-54. doi: 10.1002/jcb.22197. J Cell Biochem. 2009. PMID: 19459163
-
Ethanol-induced expression of glutamate-cysteine ligase catalytic subunit gene is mediated by NF-kappaB.Toxicol Lett. 2009 Mar 10;185(2):110-5. doi: 10.1016/j.toxlet.2008.12.006. Epub 2008 Dec 14. Toxicol Lett. 2009. PMID: 19133322
-
Effect of arsenic on transcription factor AP-1 and NF-kappaB DNA binding activity and related gene expression.Toxicol Lett. 2002 Jul 7;133(1):33-45. doi: 10.1016/s0378-4274(02)00083-8. Toxicol Lett. 2002. PMID: 12076508 Review.
-
An important role of Nrf2-ARE pathway in the cellular defense mechanism.J Biochem Mol Biol. 2004 Mar 31;37(2):139-43. doi: 10.5483/bmbrep.2004.37.2.139. J Biochem Mol Biol. 2004. PMID: 15469687 Review.
Cited by
-
In Vivo Brain Glutathione is Higher in Older Age and Correlates with Mobility.Cereb Cortex. 2021 Aug 26;31(10):4576-4594. doi: 10.1093/cercor/bhab107. Cereb Cortex. 2021. PMID: 33959751 Free PMC article.
-
Prevention of Oxygen-Induced Inflammatory Lung Injury by Caffeine in Neonatal Rats.Oxid Med Cell Longev. 2020 Aug 7;2020:3840124. doi: 10.1155/2020/3840124. eCollection 2020. Oxid Med Cell Longev. 2020. PMID: 32831996 Free PMC article.
-
Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells.PLoS One. 2012;7(12):e51111. doi: 10.1371/journal.pone.0051111. Epub 2012 Dec 7. PLoS One. 2012. PMID: 23236440 Free PMC article.
-
Potential antioxidant response to coffee - A matter of genotype?Meta Gene. 2014 Aug 7;2:525-39. doi: 10.1016/j.mgene.2014.07.003. eCollection 2014 Dec. Meta Gene. 2014. PMID: 25606436 Free PMC article.
-
Cytochrome c Deficiency Confers Apoptosome and Mitochondrial Dysfunction in African-American Men with Prostate Cancer.Cancer Res. 2019 Apr 1;79(7):1353-1368. doi: 10.1158/0008-5472.CAN-18-2383. Epub 2019 Feb 14. Cancer Res. 2019. PMID: 30765600 Free PMC article.
References
-
- Chan, J. Y., and M. Kwong. 2000. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim. Biophys. Acta 1517:19-26. - PubMed
-
- Erickson, A. M., Z. Nevarea, J. J. Gipp, and R. T. Mulcahy. 2002. Identification of a variant antioxidant response element in the promoter of the human glutamate-cysteine ligase modifier subunit gene. J. Biol. Chem. 277:30730-30737. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
