Multiple roles of vertebrate REV genes in DNA repair and recombination
- PMID: 15988022
- PMCID: PMC1168817
- DOI: 10.1128/MCB.25.14.6103-6111.2005
Multiple roles of vertebrate REV genes in DNA repair and recombination
Abstract
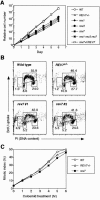
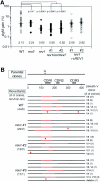
In yeast, Rev1, Rev3, and Rev7 are involved in translesion synthesis over various kinds of DNA damage and spontaneous and UV-induced mutagenesis. Here, we disrupted Rev1, Rev3, and Rev7 in the chicken B-lymphocyte line DT40. REV1-/- REV3-/- REV7-/- cells showed spontaneous cell death, chromosomal instability/fragility, and hypersensitivity to various genotoxic treatments as observed in each of the single mutants. Surprisingly, the triple-knockout cells showed a suppressed level of sister chromatid exchanges (SCEs), which may reflect postreplication repair events mediated by homologous recombination, while each single mutant showed an elevated SCE level. Furthermore, REV1-/- cells as well as triple mutants showed a decreased level of immunoglobulin gene conversion, suggesting participation of Rev1 in a recombination-based pathway. The present study gives us a new insight into cooperative function of three Rev molecules and the Polzeta (Rev3-Rev7)-independent role of Rev1 in vertebrate cells.
Figures




References
-
- Baynton, K., A. Bresson-Roy, and R. P. Fuchs. 1999. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 34:124-133. - PubMed
-
- Bemark, M., A. A. Khamlichi, S. L. Davies, and M. S. Neuberger. 2000. Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol. 10:1213-1216. - PubMed
-
- Bezzubova, O., A. Silbergleit, Y. Yamaguchi-Iwai, S. Takeda, and J. M. Buerstedde. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell 89:185-193. - PubMed
-
- Broomfield, S., T. Hryciw, and W. Xiao. 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486:167-184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
