Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis
- PMID: 15988031
- PMCID: PMC1168798
- DOI: 10.1128/MCB.25.14.6225-6234.2005
Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis
Abstract
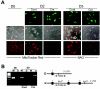
Although several genes involved in mitochondrial function are direct Myc targets, the role of Myc in mitochondrial biogenesis has not been directly established. We determined the effects of ectopic Myc expression or the loss of Myc on mitochondrial biogenesis. Induction of Myc in P493-6 cells resulted in increased oxygen consumption and mitochondrial mass and function. Conversely, compared to wild-type Myc fibroblasts, Myc null rat fibroblasts have diminished mitochondrial mass and decreased number of normal mitochondria. Reconstitution of Myc expression in Myc null fibroblasts partially restored mitochondrial mass and function and normal-appearing mitochondria. Concordantly, we also observed in primary hepatocytes that acute deletion of floxed murine Myc by Cre recombinase resulted in diminished mitochondrial mass in primary hepatocytes. Our microarray analysis of genes responsive to Myc in human P493-6 B lymphocytes supports a role for Myc in mitochondrial biogenesis, since genes involved in mitochondrial structure and function are overrepresented among the Myc-induced genes. In addition to the known direct binding of Myc to many genes involved in mitochondrial structure and function, we found that Myc binds the TFAM gene, which encodes a key transcriptional regulator and mitochondrial DNA replication factor, both in P493-6 lymphocytes with high ectopic MYC expression and in serum-stimulated primary human 2091 fibroblasts with induced endogenous MYC. These observations support a pivotal role for Myc in regulating mitochondrial biogenesis.
Figures

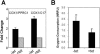



References
-
- Amati, B., K. Alevizopoulos, and J. Vlach. 1998. Myc and the cell cycle. Front Biosci. 3:d250-d268. - PubMed
-
- Arabi, A., S. Wu, K. Ridderstrale, H. Bierhoff, C. Shiue, K. Fatyol, S. Fahlen, P. Hydbring, O. Soderberg, I. Grummt, L. G. Larsson, and A. P. Wright. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell. Biol. 7:303-310. - PubMed
-
- Cam, H., E. Balciunaite, A. Blais, A. Spektor, R. C. Scarpulla, R. Young, Y. Kluger, and B. D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16:399-411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
