The oncogenic TLS-ERG fusion protein exerts different effects in hematopoietic cells and fibroblasts
- PMID: 15988032
- PMCID: PMC1168819
- DOI: 10.1128/MCB.25.14.6235-6246.2005
The oncogenic TLS-ERG fusion protein exerts different effects in hematopoietic cells and fibroblasts
Abstract
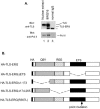
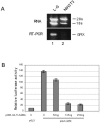
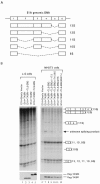
The oncogenic TLS-ERG fusion protein is found in human myeloid leukemia and Ewing's sarcoma as a result of specific chromosomal translocation. To unveil the potential mechanism(s) underlying cellular transformation, we have investigated the effects of TLS-ERG on both gene transcription and RNA splicing. Here we show that the TLS protein forms complexes with RNA polymerase II (Pol II) and the serine-arginine family of splicing factors in vivo. Deletion analysis of TLS-ERG in both mouse L-G myeloid progenitor cells and NIH 3T3 fibroblasts revealed that the RNA Pol II-interacting domain of TLS-ERG resides within the first 173 amino acids. While TLS-ERG repressed expression of the luciferase reporter gene driven by glycoprotein IX promoter in L-G cells but not in NIH 3T3 cells, the fusion protein was able to affect splicing of the E1A reporter in NIH 3T3 cells but not in L-G cells. To identify potential target genes of TLS-ERG, the fusion protein and its mutants were stably expressed in both L-G and NIH 3T3 cells through retroviral transduction. Microarray analysis of RNA samples from these cells showed that TLS-ERG activates two different sets of genes sharing little similarity in the two cell lines. Taken together, these results suggest that the oncogenic TLS-ERG fusion protein transforms hematopoietic cells and fibroblasts via different pathways.
Figures






Similar articles
-
Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein.Cancer Res. 2001 May 1;61(9):3586-90. Cancer Res. 2001. PMID: 11325824
-
TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine-arginine proteins.Mol Cell Biol. 2000 May;20(10):3345-54. doi: 10.1128/MCB.20.10.3345-3354.2000. Mol Cell Biol. 2000. PMID: 10779324 Free PMC article.
-
Dual transforming activities of the FUS (TLS)-ERG leukemia fusion protein conferred by two N-terminal domains of FUS (TLS).Mol Cell Biol. 1999 Nov;19(11):7639-50. doi: 10.1128/MCB.19.11.7639. Mol Cell Biol. 1999. PMID: 10523652 Free PMC article.
-
Rare and favorable prognosis of pediatric acute lymphoblastic leukemia with TLS-ERG fusion gene: Case report with long-term follow-up and review of literature.Cancer Genet. 2021 Aug;256-257:51-56. doi: 10.1016/j.cancergen.2021.04.003. Epub 2021 Apr 10. Cancer Genet. 2021. PMID: 33894645 Review.
-
TLS, EWS and TAF15: a model for transcriptional integration of gene expression.Brief Funct Genomic Proteomic. 2006 Mar;5(1):8-14. doi: 10.1093/bfgp/ell015. Epub 2006 Feb 23. Brief Funct Genomic Proteomic. 2006. PMID: 16769671 Review.
Cited by
-
TASR-1 regulates alternative splicing of collagen genes in chondrogenic cells.Biochem Biophys Res Commun. 2007 May 4;356(2):411-7. doi: 10.1016/j.bbrc.2007.02.159. Epub 2007 Mar 9. Biochem Biophys Res Commun. 2007. PMID: 17367759 Free PMC article.
-
Modeling initiation of Ewing sarcoma in human neural crest cells.PLoS One. 2011 Apr 29;6(4):e19305. doi: 10.1371/journal.pone.0019305. PLoS One. 2011. PMID: 21559395 Free PMC article.
-
ERG deregulation induces PIM1 over-expression and aneuploidy in prostate epithelial cells.PLoS One. 2011;6(11):e28162. doi: 10.1371/journal.pone.0028162. Epub 2011 Nov 30. PLoS One. 2011. PMID: 22140532 Free PMC article.
-
A causal role for ERG in neoplastic transformation of prostate epithelium.Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):2105-10. doi: 10.1073/pnas.0711711105. Epub 2008 Feb 1. Proc Natl Acad Sci U S A. 2008. PMID: 18245377 Free PMC article.
-
Deregulation of DUX4 and ERG in acute lymphoblastic leukemia.Nat Genet. 2016 Dec;48(12):1481-1489. doi: 10.1038/ng.3691. Epub 2016 Oct 24. Nat Genet. 2016. PMID: 27776115 Free PMC article.
References
-
- Arvand, A., H. Bastians, S. M. Welford, A. D. Thompson, J. V. Ruderman, and C. T. Denny. 1998. EWS/FLI1 up regulates mE2-C, a cyclin-selective ubiquitin conjugating enzyme involved in cyclin B destruction. Oncogene 17:2039-2045. - PubMed
-
- Bastian, L. S., M. Yagi, C. Chan, and G. J. Roth. 1996. Analysis of the megakaryocyte glycoprotein IX promoter identifies positive and negative regulatory domains and functional GATA and Ets sites. J. Biol. Chem. 271:18554-18560. - PubMed
-
- Blackburn, M. L., H. A. Chansky, A. Zielinska-Kwiatkowska, Y. Matsui, and L. Yang. 2003. Genomic structure and expression of the mouse ESET gene encoding an ERG-associated histone methyltransferase with a SET domain. Biochim. Biophys. Acta 1629:8-14. - PubMed
-
- Caceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
