A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the international consortium for prostate cancer genetics
- PMID: 15988677
- PMCID: PMC1224525
- DOI: 10.1086/432377
A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the international consortium for prostate cancer genetics
Abstract
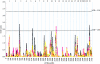
Evidence of the existence of major prostate cancer (PC)-susceptibility genes has been provided by multiple segregation analyses. Although genomewide screens have been performed in over a dozen independent studies, few chromosomal regions have been consistently identified as regions of interest. One of the major difficulties is genetic heterogeneity, possibly due to multiple, incompletely penetrant PC-susceptibility genes. In this study, we explored two approaches to overcome this difficulty, in an analysis of a large number of families with PC in the International Consortium for Prostate Cancer Genetics (ICPCG). One approach was to combine linkage data from a total of 1,233 families to increase the statistical power for detecting linkage. Using parametric (dominant and recessive) and nonparametric analyses, we identified five regions with "suggestive" linkage (LOD score >1.86): 5q12, 8p21, 15q11, 17q21, and 22q12. The second approach was to focus on subsets of families that are more likely to segregate highly penetrant mutations, including families with large numbers of affected individuals or early age at diagnosis. Stronger evidence of linkage in several regions was identified, including a "significant" linkage at 22q12, with a LOD score of 3.57, and five suggestive linkages (1q25, 8q13, 13q14, 16p13, and 17q21) in 269 families with at least five affected members. In addition, four additional suggestive linkages (3p24, 5q35, 11q22, and Xq12) were found in 606 families with mean age at diagnosis of < or = 65 years. Although it is difficult to determine the true statistical significance of these findings, a conservative interpretation of these results would be that if major PC-susceptibility genes do exist, they are most likely located in the regions generating suggestive or significant linkage signals in this large study.
Figures



References
Web Resource
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PC, ELAC2, RNASEL, MSR1, HPC1, and CHEK2)
References
Publication types
MeSH terms
Substances
Grants and funding
- CA89600/CA/NCI NIH HHS/United States
- N01 HG065403/HG/NHGRI NIH HHS/United States
- N01-PC-35141/PC/NCI NIH HHS/United States
- CA72818/CA/NCI NIH HHS/United States
- CA67044/CA/NCI NIH HHS/United States
- CA080122/CA/NCI NIH HHS/United States
- R01 CA106523/CA/NCI NIH HHS/United States
- R01 CA067044/CA/NCI NIH HHS/United States
- T32 CA080416/CA/NCI NIH HHS/United States
- CA079596/CA/NCI NIH HHS/United States
- M01-RR00064/RR/NCRR NIH HHS/United States
- R01 CA090752/CA/NCI NIH HHS/United States
- R01 CA89600/CA/NCI NIH HHS/United States
- P50 CA058236/CA/NCI NIH HHS/United States
- R01 CA095052/CA/NCI NIH HHS/United States
- U01 CA089600/CA/NCI NIH HHS/United States
- CA78835/CA/NCI NIH HHS/United States
- U01 CA067044/CA/NCI NIH HHS/United States
- CA58236/CA/NCI NIH HHS/United States
- CA106523-01A1/CA/NCI NIH HHS/United States
- R01 CA072818/CA/NCI NIH HHS/United States
- CA95052-01/CA/NCI NIH HHS/United States
- K07 CA098364/CA/NCI NIH HHS/United States
- R01 CA079596/CA/NCI NIH HHS/United States
- R01 CA90752/CA/NCI NIH HHS/United States
- M01 RR000064/RR/NCRR NIH HHS/United States
- N01 PC035141/CA/NCI NIH HHS/United States
- R01 CA080122/CA/NCI NIH HHS/United States
- K07 CA98364/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

