Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human mitochondrial NAD(P)+-dependent malate dehydrogenase (malic enzyme)
- PMID: 15989682
- PMCID: PMC1317662
- DOI: 10.1042/BJ20050641
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human mitochondrial NAD(P)+-dependent malate dehydrogenase (malic enzyme)
Abstract
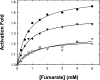
Human mitochondrial NAD(P)+-dependent malate dehydrogenase (decarboxylating) (malic enzyme) can be specifically and allosterically activated by fumarate. X-ray crystal structures have revealed conformational changes in the enzyme in the absence and in the presence of fumarate. Previous studies have indicated that fumarate is bound to the allosteric pocket via Arg67 and Arg91. Mutation of these residues almost abolishes the activating effect of fumarate. However, these amino acid residues are conserved in some enzymes that are not activated by fumarate, suggesting that there may be additional factors controlling the activation mechanism. In the present study, we tried to delineate the detailed molecular mechanism of activation of the enzyme by fumarate. Site-directed mutagenesis was used to replace Asp102, which is one of the charged amino acids in the fumarate binding pocket and is not conserved in other decarboxylating malate dehydrogenases. In order to explore the charge effect of this residue, Asp102 was replaced by alanine, glutamate or lysine. Our experimental data clearly indicate the importance of Asp102 for activation by fumarate. Mutation of Asp102 to Ala or Lys significantly attenuated the activating effect of fumarate on the enzyme. Kinetic parameters indicate that the effect of fumarate was mainly to decrease the K(m) values for malate, Mg2+ and NAD+, but it did not notably elevate kcat. The apparent substrate K(m) values were reduced by increasing concentrations of fumarate. Furthermore, the greatest effect of fumarate activation was apparent at low malate, Mg2+ or NAD+ concentrations. The K(act) values were reduced with increasing concentrations of malate, Mg2+ and NAD+. The Asp102 mutants, however, are much less sensitive to regulation by fumarate. Mutation of Asp102 leads to the desensitization of the co-operative effect between fumarate and substrates of the enzyme.
Figures




Similar articles
-
Long-range interaction between the enzyme active site and a distant allosteric site in the human mitochondrial NAD(P)+-dependent malic enzyme.Arch Biochem Biophys. 2009 Jul 1;487(1):19-27. doi: 10.1016/j.abb.2009.05.007. Epub 2009 May 22. Arch Biochem Biophys. 2009. PMID: 19464998
-
Ascaris suum NAD-malic enzyme is activated by L-malate and fumarate binding to separate allosteric sites.Biochemistry. 2003 Aug 19;42(32):9712-21. doi: 10.1021/bi034101w. Biochemistry. 2003. PMID: 12911313
-
Dual roles of Lys(57) at the dimer interface of human mitochondrial NAD(P)+-dependent malic enzyme.Biochem J. 2009 May 13;420(2):201-9. doi: 10.1042/BJ20090076. Biochem J. 2009. PMID: 19236308
-
Malate dehydrogenase: a model for structure, evolution, and catalysis.Protein Sci. 1994 Oct;3(10):1883-8. doi: 10.1002/pro.5560031027. Protein Sci. 1994. PMID: 7849603 Free PMC article. Review.
-
Catalytic mechanism and kinetics of malate dehydrogenase.Essays Biochem. 2024 Oct 3;68(2):73-82. doi: 10.1042/EBC20230086. Essays Biochem. 2024. PMID: 38721782 Free PMC article. Review.
Cited by
-
Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.Heliyon. 2022 Dec 19;8(12):e12392. doi: 10.1016/j.heliyon.2022.e12392. eCollection 2022 Dec. Heliyon. 2022. PMID: 36590518 Free PMC article.
-
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties.Plant Mol Biol. 2008 Jun;67(3):231-42. doi: 10.1007/s11103-008-9313-9. Plant Mol Biol. 2008. PMID: 18288573
-
Structural insights into the allosteric site of Arabidopsis NADP-malic enzyme 2: role of the second sphere residues in the regulatory signal transmission.Plant Mol Biol. 2021 Sep;107(1-2):37-48. doi: 10.1007/s11103-021-01176-2. Epub 2021 Jul 31. Plant Mol Biol. 2021. PMID: 34333694
-
Metal ions stabilize a dimeric molten globule state between the open and closed forms of malic enzyme.Biophys J. 2007 Dec 1;93(11):3977-88. doi: 10.1529/biophysj.107.111385. Epub 2007 Aug 17. Biophys J. 2007. PMID: 17704184 Free PMC article.
-
Functional roles of the tetramer organization of malic enzyme.J Biol Chem. 2009 Jul 3;284(27):18096-105. doi: 10.1074/jbc.M109.005082. Epub 2009 May 5. J Biol Chem. 2009. PMID: 19416979 Free PMC article.
References
-
- Frenkel R. Regulation and physiological function of malic enzyme. Curr. Top. Cell. Regul. 1975;9:157–181. - PubMed
-
- Hsu R. Y. Pigeon liver malic enzyme. Mol. Cell. Biochem. 1982;43:3–26. - PubMed
-
- Loeber G., Infante A. A., Maurer-Fogy I., Krystek E., Dworkin M. B. Human NAD+-dependent mitochondrial malic enzyme. J. Biol. Chem. 1991;266:3016–3021. - PubMed
-
- Rao G. S. J., Coleman D. E., Kulkarni G., Goldsmith E. J., Cook P. F., Harris B. G. NAD-malic enzyme from Ascaris suum: sequence and structural studies. Protein Pept. Lett. 2000;7:297–304.
-
- Cleland W. W. Chemical mechanism of malic enzyme as determined by isotope effects and alternate substrates. Protein Pept. Lett. 2000;7:305–312.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

