The MAP kinase substrate MKS1 is a regulator of plant defense responses
- PMID: 15990873
- PMCID: PMC1176463
- DOI: 10.1038/sj.emboj.7600737
The MAP kinase substrate MKS1 is a regulator of plant defense responses
Abstract
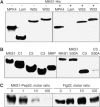
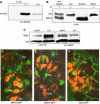
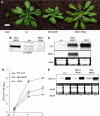
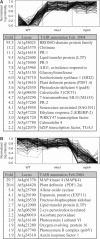
Arabidopsis MAP kinase 4 (MPK4) functions as a regulator of pathogen defense responses, because it is required for both repression of salicylic acid (SA)-dependent resistance and for activation of jasmonate (JA)-dependent defense gene expression. To understand MPK4 signaling mechanisms, we used yeast two-hybrid screening to identify the MPK4 substrate MKS1. Analyses of transgenic plants and genome-wide transcript profiling indicated that MKS1 is required for full SA-dependent resistance in mpk4 mutants, and that overexpression of MKS1 in wild-type plants is sufficient to activate SA-dependent resistance, but does not interfere with induction of a defense gene by JA. Further yeast two-hybrid screening revealed that MKS1 interacts with the WRKY transcription factors WRKY25 and WRKY33. WRKY25 and WRKY33 were shown to be in vitro substrates of MPK4, and a wrky33 knockout mutant was found to exhibit increased expression of the SA-related defense gene PR1. MKS1 may therefore contribute to MPK4-regulated defense activation by coupling the kinase to specific WRKY transcription factors.
Figures


References
-
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 - PubMed
-
- Baker DA, Mille-Baker B, Wainwright SM, Ish-Horowicz D, Dibb NJ (2001) Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature 411: 330–334 - PubMed
-
- Bao MZ, Schwartz MA, Cantin GT, Yates JR III, Madhani HD (2004) Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119: 991–1000 - PubMed
-
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

