Comparison of 9 different PCR primers for the rapid detection of severe acute respiratory syndrome coronavirus using 2 RNA extraction methods
- PMID: 15994050
- PMCID: PMC7125917
- DOI: 10.1016/j.diagmicrobio.2005.03.007
Comparison of 9 different PCR primers for the rapid detection of severe acute respiratory syndrome coronavirus using 2 RNA extraction methods
Abstract
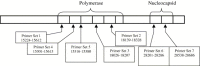
The sensitivity and specificity of various severe acute respiratory syndrome coronavirus (SARS-CoV) PCR primer and probe sets were evaluated through the use of commercial kits and in-house amplification formats. Conventional and real-time PCR assays were performed using a heat-block thermocycler ABI 9600, the Roche LightCycler version 1.2, or the ABI 7000 Sequence Detection System. The sensitivity of all primers was between 0.0004 and 0.04 PFU with viral cell lysate and between 0.004 and 0.4 PFU in spiked stool specimen per PCR assay. The primer sets for real-time PCR assays were at one least 1 log more sensitive than the primer sets used in the conventional PCR. A panel of viruses including swine gastroenteritis virus, bovine coronavirus, avian bronchitis virus (Connecticut strain), avian bronchitis virus (Massachusetts strain), human coronaviruses 229E and OC43, parainfluenza virus (type III), human metapneumovirus, adenovirus, respiratory syncytial virus, and influenza A were tested by all assays. All real-time PCR assays used probe-based detection, and no cross-reactivity was observed. With conventional PCR, analysis was performed using agarose gel electrophoresis and multiple nonspecific bands were observed. Two commercial extraction methods, magnetic particle capture and silica-based procedure were evaluated and the results were comparable. The former was less laborious with shorter time for completion and can easily be adapted to an automated system such as the MagNa Pure-LC, which can extract nucleic acid from clinical samples and load it into the sample capillaries of the LightCycler. As exemplified by this study, the continued refinement and evaluation of PCR procedures will greatly benefit the diagnostic laboratory during an outbreak of SARS.
Figures


References
-
- Cusi M.G., Valassina M., Valensin P.E. Comparison of M-MLV reverse transcriptase and Tth polymerase activity in RT-PCR of samples with low virus burden. Biotechniques. 1994;17:1034–1036. - PubMed
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Drosten C., Chiu L.L., Panning M., Leong H.N., Preiser W., Tam J.S., Gunther S., Kramme S., Emmerich P., Ng W.L., Schmitz H., Koay E. Evaluation of advanced reverse transcription-PCR assays and an alternative PCR target region for detection of severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol. 2004;42:2043–2047. - PMC - PubMed
-
- Emery S.L., Erdman D.D., Bowen M.D., Newton B.R., Winchell J.M., Meyer R.F., Tong S., Cook B.T., Holloway B.P, McCaustland K.A., Rota P.A., Bankamp B., Lowe L.E., Ksiazek T.G., Bellini W.J., Anderson L.J. Real-time polymerase chain reaction for detecting SARS coronavirus, Beijing, 2003. Emerg. Infect. Dis. 2004;10:311–313. - PMC - PubMed
-
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo W.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yune K.Y., Peiris J.S.M., Poon L.L.M. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

