Single-molecule studies of repressor-DNA interactions show long-range interactions
- PMID: 15994229
- PMCID: PMC1168954
- DOI: 10.1073/pnas.0502917102
Single-molecule studies of repressor-DNA interactions show long-range interactions
Abstract
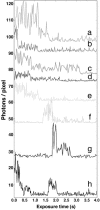
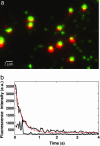
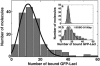
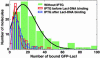
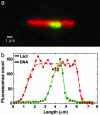
We have performed single-molecule studies of GFP-LacI repressor proteins bound to bacteriophage lambda DNA containing a 256 tandem lac operator insertion confined in nanochannels. An integrated photon molecular counting method was developed to determine the number of proteins bound to DNA. By using this method, we determined the saturated mean occupancy of the 256 tandem lac operators to be 13, which constitutes only 2.5% of the available sites. This low occupancy level suggests that the repressors influence each other even when they are widely separated, at distances on the order of 200 nm, or several DNA persistence lengths.
Figures






References
-
- Jacob, F. & Monod, J. J. (1961) J. Mol. Biol. 3, 318–356. - PubMed
-
- Lewis, M., Chan, G., Horton, N. C., Kercher, M. A., Pace, H. C., Schumacher, M. A., Brennan, R. G. & Lu, P. (1996) Science 271, 1247–1254. - PubMed
-
- Kalodimos, C. G., Biris, N, Bonvin, A. M. J. J., Levandoski, M. M., Guennuegues, M., Boelens, R. & Kaptein, R. (2004) Science 305, 386–389. - PubMed
-
- Bell, C. E. & Lewis, M. (2000) Nat. Struct. Biol. 7, 209–214. - PubMed
-
- Levandoski, M. M., Tsodikov, O. V., Frank, D. E., Melcher, S. E., Saecker, R. M. & Record, M. T., Jr. (1996) J. Mol. Biol. 260, 697–717. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

