Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1
- PMID: 15994775
- PMCID: PMC1168742
- DOI: 10.1128/JVI.79.14.8812-8827.2005
Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1
Abstract
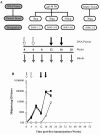
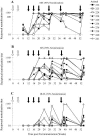
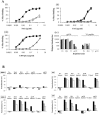
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) complex comprises three gp120 exterior glycoproteins each noncovalently linked to a gp41 transmembrane glycoprotein. Monomeric gp120 proteins can elicit antibodies capable of neutralizing atypically sensitive test viruses in vitro, but these antibodies are ineffective against representative primary isolates and the gp120 vaccines failed to provide protection against HIV-1 transmission in vivo. Alternative approaches to raising neutralizing antibodies are therefore being pursued. Here we report on the antibody responses generated in rabbits against a soluble, cleaved, trimeric form of HIV-1(JR-FL) Env. In this construct, the gp120 and gp41 moieties are covalently linked by an intermolecular disulfide bond (SOS gp140), and an I559P substitution has been added to stabilize gp41-gp41 interactions (SOSIP gp140). We investigated the value of DNA priming and compared the use of membrane-bound and soluble priming antigens and of repeat boosting with soluble and particulate protein antigen. Compared to monomeric gp120, SOSIP gp140 trimers elicited approximately threefold lower titers of anti-gp120 antibodies. Priming with DNA encoding a membrane-bound form of the SOS gp140 protein, followed by several immunizations with soluble SOSIP gp140 trimers, resulted in antibodies capable of neutralizing sensitive strains at high titers. A subset of these sera also neutralized, at lower titers, HIV-1(JR-FL) and some other primary isolates in pseudovirus and/or whole-virus assays. Neutralization of these viruses was immunoglobulin mediated and was predominantly caused by antibodies to gp120 epitopes, but not the V3 region.
Figures


References
-
- Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. - PMC - PubMed
-
- Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. - PubMed
-
- Binley, J. M., R. W. Sanders, B. Clas, N. Schulke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

