Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm
- PMID: 15994793
- PMCID: PMC1168739
- DOI: 10.1128/JVI.79.14.8991-9005.2005
Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm
Abstract
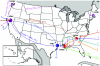
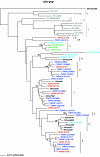
Retrospective molecular epidemiology was performed on samples from four sooty mangabey (SM) colonies in the United States to characterize simian immunodeficiency virus SIVsm diversity in SMs and to trace virus circulation among different primate centers (PCs) over the past 30 years. The following SIVsm sequences were collected from different monkeys: 55 SIVsm isolates from the Tulane PC sampled between 1984 and 2004, 10 SIVsm isolates from the Yerkes PC sampled in 2002, 7 SIVsm isolates from the New Iberia PC sampled between 1979 and 1986, and 8 SIVsm isolates from the California PC sampled between 1975 and 1977. PCR and sequencing were done to characterize the gag, pol, and env gp36 genes. Phylogenetic analyses were correlated with the epidemiological data. Our analysis identified nine different divergent phylogenetic lineages that cocirculated in these four SM colonies in the Unites States in the past 30 years. Lineages 1 to 5 have been identified previously. Two of the newly identified SIVsm lineages found in SMs are ancestral to SIVmac251/SIVmac239/SIVmne and SIVstm. We further identified the origin of these two macaque viruses in SMs from the California National Primate Research Center. The diversity of SIVsm isolates in PCs in the United States mirrors that of human immunodeficiency virus type 1 (HIV-1) group M subtypes and offers a model for the molecular epidemiology of HIV and a new approach to vaccine testing. The cocirculation of divergent SIVsm strains in PCs resulted in founder effects, superinfections, and recombinations. This large array of SIVsm strains showing the same magnitude of diversity as HIV-1 group M subtypes should be extremely useful for modeling the efficacy of vaccination strategies under the real-world conditions of HIV-1 diversity. The genetic variability of SIVsm strains among PCs may influence the diagnosis and monitoring of SIVsm infection and, consequently, may bias the results of pathogenesis studies.
Figures




References
-
- Apetrei, C., B. Gormus, I. Pandrea, M. Metzger, P. ten Haaft, L. N. Martin, R. Bohm, X. Alvarez, G. Koopman, M. Murphey-Corb, R. S. Veazey, A. A. Lackner, G. Baskin, J. Heeney, and P. A. Marx. 2004. Direct inoculation of simian immunodeficiency virus from sooty mangabeys in black mangabeys (Lophocebus aterrimus): first evidence of AIDS in a heterologous African species and different pathologic outcomes of experimental infection. J. Virol. 78:11506-11518. - PMC - PubMed
-
- Apetrei, C., P. A. Marx, and S. M. Smith. 2004. The evolution of HIV and its consequences. Infect. Dis. Clin. N. Am. 18:369-394. - PubMed
-
- Apetrei, C., A. Necula, C. Holm-Hansen, I. Loussert-Ajaka, I. Pandrea, C. Cozmei, A. Streinu-Cercel, F. R. Pascu, E. Negut, G. Molnar, M. Duca, M. Pecec, F. Brun-Vezinet, and F. Simon. 1998. HIV-1 diversity in Romania. AIDS 12:1079-1085. - PubMed
-
- Apetrei, C., D. L. Robertson, and P. A. Marx. 2004. The history of SIVs and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front. Biosci. 9:225-254. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

