Compartmentalization of hepatitis C virus quasispecies in blood mononuclear cells of patients with mixed cryoglobulinemic syndrome
- PMID: 15994809
- PMCID: PMC1168762
- DOI: 10.1128/JVI.79.14.9145-9156.2005
Compartmentalization of hepatitis C virus quasispecies in blood mononuclear cells of patients with mixed cryoglobulinemic syndrome
Abstract
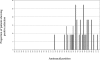
The aim of this study was to investigate the quasispecies heterogeneity of hepatitis C virus (HCV) in the plasma, cryoprecipitate, and peripheral lymphocytes of chronically infected HCV patients with mixed cryoglobulinemia (MC). We studied 360 clones from 10 HCV-positive patients with MC and 8 age-, gender- and HCV genotype-matched subjects with chronic HCV infection but without MC. A partial nucleotide sequence encompassing the E1/E2 region, including hypervariable region 1 (HVR1), was amplified and cloned from plasma, cryoprecipitates, and peripheral blood mononuclear cells (PBMC), and the genetic diversity and complexity and synonymous and nonsynonymous substitution rates were determined. Heterogeneous selection pressure at codon sites was evaluated. Compartmentalization was estimated by phylogenetic and phenetic (Mantel's test) approaches. The patients with MC had 3.3 times lower nonsynonymous substitution rates (1.7 versus 5.7 substitutions/100 sites). Among the subjects with HCV genotype 1, the MC patients had significantly less complexity than the controls, whereas the diversity and complexity were similar in the genotype 2 patients and controls. Site-specific selection analysis confirmed the low frequency of MC patients showing positive selection. There was a significant correlation between positive selection and the infecting HCV genotype. The quasispecies were less heterogeneous in PBMC than in plasma. Significant compartmentalization of HCV quasispecies was observed in the PBMC of four of nine subjects (three with MC) and seven of nine cryoprecipitates. In one subject with MC, we detected a 5-amino-acid insertion at codons 385 to 389 of HVR1. Our results suggest reduced quasispecies heterogeneity in MC patients that is related to a low selection pressure which is probably due to an impaired immune response, the HCV genotype, and/or the duration of the infection. The frequent HCV quasispecies compartmentalization in patients' PBMC suggests a possible pathogenetic significance.
Figures







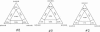
References
-
- Agnello, V., R. T. Chung, and L. M. Kaplan. 1992. A role for hepatitis C virus infection in type II cryoglobulinemia. N. Engl. J. Med. 327:1490-1495. - PubMed
-
- Aiyama, T., K. Yoshioka, A. Okumura, M. Takayanagi, K. Iwata, T. Ishikawa, and S. Kakumu. 1996. Hypervariable region sequence in cryoglobulin-associated hepatitis C virus in sera of patients with chronic hepatitis C: relationship to antibody response against hypervariable region genome. Hepatology 24:1346-1350. - PubMed
-
- Asselah, T., M. Martinot, D. Cazals-Hatem, N. Boyer, A. Auperin, V. Le Breton, S. Erlinger, C. Degott, D. Valla, and P. Marcellin. 2002. Hypervariable region 1 quasispecies in hepatitis C virus genotypes 1b and 3 infected patients with normal and abnormal alanine aminotransferase levels. J. Viral Hepat. 9:29-35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

