The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains
- PMID: 15995192
- PMCID: PMC1169503
- DOI: 10.1128/JB.187.14.4774-4781.2005
The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains
Abstract
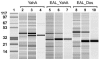
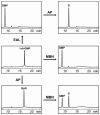
The EAL domain (also known as domain of unknown function 2 or DUF2) is a ubiquitous signal transduction protein domain in the Bacteria. Its involvement in hydrolysis of the novel second messenger cyclic dimeric GMP (c-di-GMP) was demonstrated in vivo but not in vitro. The EAL domain-containing protein Dos from Escherichia coli was reported to hydrolyze cyclic AMP (cAMP), implying that EAL domains have different substrate specificities. To investigate the biochemical activity of EAL, the E. coli EAL domain-containing protein YahA and its individual EAL domain were overexpressed, purified, and characterized in vitro. Both full-length YahA and the EAL domain hydrolyzed c-di-GMP into linear dimeric GMP, providing the first biochemical evidence that the EAL domain is sufficient for phosphodiesterase activity. This activity was c-di-GMP specific, optimal at alkaline pH, dependent on Mg(2+) or Mn(2+), strongly inhibited by Ca(2+), and independent of protein oligomerization. Linear dimeric GMP was shown to be 5'pGpG. The EAL domain from Dos was overexpressed, purified, and found to function as a c-di-GMP-specific phosphodiesterase, not as a cAMP-specific phosphodiesterase, in contrast to previous reports. The EAL domains can hydrolyze 5'pGpG into GMP, however, very slowly, thus implying that this activity is irrelevant in vivo. Therefore, c-di-GMP is the exclusive substrate of EAL. Multiple-sequence alignment revealed two groups of EAL domains hypothesized to correspond to enzymatically active and inactive domains. The domains in the latter group have mutations in residues conserved in the active domains. The enzymatic inactivity of EAL domains may explain their coexistence with GGDEF domains in proteins possessing c-di-GMP synthase (diguanulate cyclase) activity.
Figures




References
-
- Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. - PubMed
-
- Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

