Synthetic biology
- PMID: 15995697
- PMCID: PMC7097405
- DOI: 10.1038/nrg1637
Synthetic biology
Abstract
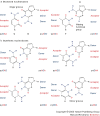
Synthetic biologists come in two broad classes. One uses unnatural molecules to reproduce emergent behaviours from natural biology, with the goal of creating artificial life. The other seeks interchangeable parts from natural biology to assemble into systems that function unnaturally. Either way, a synthetic goal forces scientists to cross uncharted ground to encounter and solve problems that are not easily encountered through analysis. This drives the emergence of new paradigms in ways that analysis cannot easily do. Synthetic biology has generated diagnostic tools that improve the care of patients with infectious diseases, as well as devices that oscillate, creep and play tic-tac-toe.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Hobom B. Surgery of genes. At the doorstep of synthetic biology. Medizin. Klinik. 1980;75:14–21. - PubMed
-
- Rawls, R. 'Synthetic Biology' makes its debut. Chem. Eng. News 49–53 (24 April 2000).
-
- Benner SA. Redesigning life. Organic chemistry and the evolution of protein. Chimia. 1987;41:142–148.
-
- Benner, S. A. Act natural. Nature421, 118 (2003). This paper outlines the importance of pursuing the science of synthetic biology. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

