Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species
- PMID: 15998412
- PMCID: PMC2442926
- DOI: 10.1111/j.1469-8137.2005.01433.x
Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species
Abstract
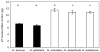
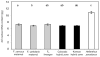
Hybridization and polyploidy can induce rapid genomic changes, including the gain or loss of DNA, but the magnitude and timing of such changes are not well understood. The homoploid hybrid system in Helianthus (three hybrid-derived species and their two parents) provides an opportunity to examine the link between hybridization and genome size changes in a replicated fashion. Flow cytometry was used to estimate the nuclear DNA content in multiple populations of three homoploid hybrid Helianthus species (Helianthus anomalus, Helianthus deserticola, and Helianthus paradoxus), the parental species (Helianthus annuus and Helianthus petiolaris), synthetic hybrids, and natural hybrid-zone populations. Results confirm that hybrid-derived species have 50% more nuclear DNA than the parental species. Despite multiple origins, hybrid species were largely consistent in their DNA content across populations, although H. deserticola showed significant interpopulation differences. First- and sixth-generation synthetic hybrids and hybrid-zone plants did not show an increase from parental DNA content. First-generation hybrids differed in DNA content according to the maternal parent. In summary, hybridization by itself does not lead to increased nuclear DNA content in Helianthus, and the evolutionary forces responsible for the repeated increases in DNA content seen in the hybrid-derived species remain mysterious.
Copyright New Phytologist (2005).
Figures

References
-
- Baranyi M, Greilhuber J. Genome size in Allium: In quest of reproducible data. Annals of Botany. 1999;83:687–695.
-
- Baumel A, Ainouche M, Kalendar R, Schulman A. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica CE Hubbard (Poaceae) Molecular Biology and Evolution. 2002;19:1218–1227. - PubMed
-
- Bennett MD, Bhandol P, Leitch IJ. Nuclear DNA amounts in angiosperms and their modern uses: 807 new estimates. Annals of Botany (London) 2000;86:859–909.
-
- Bennett MD, Leitch IJ. Plant DNA C-values database, release 2.0. 2003. http://www.kew.org/cval.
-
- Bennetzen JL. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica. 2002;115:29–36. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

