Validation and refinement of gene-regulatory pathways on a network of physical interactions
- PMID: 15998451
- PMCID: PMC1175993
- DOI: 10.1186/gb-2005-6-7-r62
Validation and refinement of gene-regulatory pathways on a network of physical interactions
Abstract
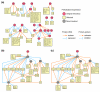
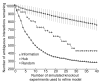
As genome-scale measurements lead to increasingly complex models of gene regulation, systematic approaches are needed to validate and refine these models. Towards this goal, we describe an automated procedure for prioritizing genetic perturbations in order to discriminate optimally between alternative models of a gene-regulatory network. Using this procedure, we evaluate 38 candidate regulatory networks in yeast and perform four high-priority gene knockout experiments. The refined networks support previously unknown regulatory mechanisms downstream of SOK2 and SWI4.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

