Efficient assembly of de novo human artificial chromosomes from large genomic loci
- PMID: 15998466
- PMCID: PMC1182356
- DOI: 10.1186/1472-6750-5-21
Efficient assembly of de novo human artificial chromosomes from large genomic loci
Abstract
Background: Human artificial chromosomes (HACs) are potentially useful vectors for gene transfer studies and for functional annotation of the genome because of their suitability for cloning, manipulating and transferring large segments of the genome. However, development of HACs for the transfer of large genomic loci into mammalian cells has been limited by difficulties in manipulating high-molecular weight DNA, as well as by the low overall frequencies of de novo HAC formation. Indeed, to date, only a small number of large (>100 kb) genomic loci have been reported to be successfully packaged into de novo HACs.
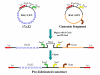
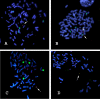
Results: We have developed novel methodologies to enable efficient assembly of HAC vectors containing any genomic locus of interest. We report here the creation of a novel, bimolecular system based on bacterial artificial chromosomes (BACs) for the construction of HACs incorporating any defined genomic region. We have utilized this vector system to rapidly design, construct and validate multiple de novo HACs containing large (100-200 kb) genomic loci including therapeutically significant genes for human growth hormone (HGH), polycystic kidney disease (PKD1) and beta-globin. We report significant differences in the ability of different genomic loci to support de novo HAC formation, suggesting possible effects of cis-acting genomic elements. Finally, as a proof of principle, we have observed sustained beta-globin gene expression from HACs incorporating the entire 200 kb beta-globin genomic locus for over 90 days in the absence of selection.
Conclusion: Taken together, these results are significant for the development of HAC vector technology, as they enable high-throughput assembly and functional validation of HACs containing any large genomic locus. We have evaluated the impact of different genomic loci on the frequency of HAC formation and identified segments of genomic DNA that appear to facilitate de novo HAC formation. These genomic loci may be useful for identifying discrete functional elements that may be incorporated into future generations of HAC vectors.
Figures


Similar articles
-
Escherichia coli-cloned CFTR loci relevant for human artificial chromosome therapy.Hum Gene Ther. 2010 Sep;21(9):1077-92. doi: 10.1089/hum.2009.225. Hum Gene Ther. 2010. PMID: 20384480
-
Construction of human artificial chromosome vectors by recombineering.Gene. 2005 May 23;351:29-38. doi: 10.1016/j.gene.2005.01.017. Epub 2005 Apr 15. Gene. 2005. PMID: 15837432
-
Human artificial chromosomes containing chromosome 17 alphoid DNA maintain an active centromere in murine cells but are not stable.Genomics. 2004 May;83(5):844-51. doi: 10.1016/j.ygeno.2003.11.011. Genomics. 2004. PMID: 15081114
-
[Research progress in human artificial chromosomes(HACs) and the potentials in application].Yi Chuan. 2005 Nov;27(6):995-1000. Yi Chuan. 2005. PMID: 16378952 Review. Chinese.
-
Advances in human artificial chromosome technology.Trends Genet. 2002 Jun;18(6):313-9. doi: 10.1016/S0168-9525(02)02679-3. Trends Genet. 2002. PMID: 12044361 Review.
Cited by
-
Generation of a conditionally self-eliminating HAC gene delivery vector through incorporation of a tTAVP64 expression cassette.Nucleic Acids Res. 2015 May 19;43(9):e57. doi: 10.1093/nar/gkv124. Epub 2015 Feb 20. Nucleic Acids Res. 2015. PMID: 25712097 Free PMC article.
-
Technique of laser chromosome welding for chromosome repair and artificial chromosome creation.Biomed Opt Express. 2018 Mar 21;9(4):1783-1794. doi: 10.1364/BOE.9.001783. eCollection 2018 Apr 1. Biomed Opt Express. 2018. PMID: 29675319 Free PMC article.
-
Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere.ACS Synth Biol. 2012 Dec 21;1(12):590-601. doi: 10.1021/sb3000436. ACS Synth Biol. 2012. PMID: 23411994 Free PMC article.
-
Human artificial chromosome vectors meet stem cells: new prospects for gene delivery.Stem Cell Rev. 2006;2(1):43-50. doi: 10.1007/s12015-006-0008-9. Stem Cell Rev. 2006. PMID: 17142886 Review.
-
Novel method to load multiple genes onto a mammalian artificial chromosome.PLoS One. 2014 Jan 15;9(1):e85565. doi: 10.1371/journal.pone.0085565. eCollection 2014. PLoS One. 2014. PMID: 24454889 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

