Proteomic analysis of a eukaryotic cilium
- PMID: 15998802
- PMCID: PMC2171396
- DOI: 10.1083/jcb.200504008
Proteomic analysis of a eukaryotic cilium
Abstract
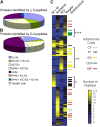
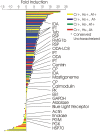
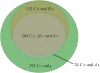
Cilia and flagella are widespread cell organelles that have been highly conserved throughout evolution and play important roles in motility, sensory perception, and the life cycles of eukaryotes ranging from protists to humans. Despite the ubiquity and importance of these organelles, their composition is not well known. Here we use mass spectrometry to identify proteins in purified flagella from the green alga Chlamydomonas reinhardtii. 360 proteins were identified with high confidence, and 292 more with moderate confidence. 97 out of 101 previously known flagellar proteins were found, indicating that this is a very complete dataset. The flagellar proteome is rich in motor and signal transduction components, and contains numerous proteins with homologues associated with diseases such as cystic kidney disease, male sterility, and hydrocephalus in humans and model vertebrates. The flagellum also contains many proteins that are conserved in humans but have not been previously characterized in any organism. The results indicate that flagella are far more complex than previously estimated.
Figures



References
-
- Avidor-Reiss, T., A.M. Maer, E. Koundakjian, A. Polyanovsky, T. Keil, S. Subramaniam, and C.S. Zuker. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 117:527–539. - PubMed
-
- Bloodgood, R.A. 1992. Calcium-regulated phosphorylation of proteins in the membrane-matrix compartment of the Chlamydomonas flagellum. Exp. Cell Res. 198:228–236. - PubMed
-
- Bradley, B.A., J.J. Wagner, and L.M. Quarmby. 2004. Identification and sequence analysis of six new members of the NIMA-related kinase family in Chlamydomonas. J. Eukaryot. Microbiol. 51:66–72. - PubMed
-
- Chae, T.H., S. Kim, K.E. Marz, P.I. Hanson, and C.A. Walsh. 2004. The hyh mutation uncovers roles for alpha Snap in apical protein localization and control of neural cell fate. Nat. Genet. 36:264–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

