The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation
- PMID: 15998807
- PMCID: PMC1172060
- DOI: 10.1101/gad.1318705
The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation
Abstract
The phytohormone abscisic acid (ABA) mediates many complex aspects of plant development including seed maturation, dormancy, and germination as well as root growth. The B3-domain transcription factor abscisic acid-insensitive 3 (ABI3) is a central regulator in ABA signaling, but little is known of how this factor is regulated. Here, we show that ABI3 is an unstable protein and that an ABI3-interacting protein (AIP2), which contains a RING motif, can polyubiquitinate ABI3 in vitro. The AIP2 E3 ligase activity is abolished by mutations (C230S; C231S) in the RING motif and the AIP2 (C/S) mutant functions in a dominant-negative manner. AIP2 has a stronger binding affinity for the B2 + B3 domain of ABI3 than the A1 + B1 domain, but only ubiquitinates the latter. In double-transgenic plants, induced AIP2 expression leads to a decrease in ABI3 protein levels. In contrast, ABI3 levels are elevated upon induced expression of the AIP2 RING mutant, which interferes with the endogenous AIP2 E3 activity. An aip2-1-null mutant shows higher ABI3 protein levels compared with wild type after seed stratification, and is hypersensitive to ABA, mimicking the ABI3-overexpression phenotype, whereas AIP2-overexpression plants contain lower levels of ABI3 protein than wild type and are more resistant to ABA, phenocopying abi3. Our results indicate that AIP2 negatively regulates ABA signaling by targeting ABI3 for post-translational destruction.
Figures






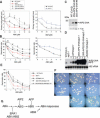
References
-
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H.M., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide Insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. - PubMed
-
- Bechtold N., Ellis, J., and Pelletier, G. 1993. In planta Agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. III 316: 1194–1199. - PubMed
-
- Beeckman T. 1994. An easy technique for clearing of histochemically stained plant tissue. Plant Mol. Biol. Rep. 82: 259–266.
-
- Brady S.M., Sarkar, S.F., Bonetta, D., and McCourt., P. 2003. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34: 67–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
