Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain
- PMID: 16000821
- PMCID: PMC1169050
- DOI: 10.1128/AEM.71.7.4057-4068.2005
Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain
Abstract
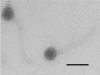
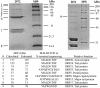
The Streptococcus thermophilus virulent pac-type phage 2972 was isolated from a yogurt made in France in 1999. It is a representative of several phages that have emerged with the industrial use of the exopolysaccharide-producing S. thermophilus strain RD534. The genome of phage 2972 has 34,704 bp with an overall G+C content of 40.15%, making it the shortest S. thermophilus phage genome analyzed so far. Forty-four open reading frames (ORFs) encoding putative proteins of 40 or more amino acids were identified, and bioinformatic analyses led to the assignment of putative functions to 23 ORFs. Comparative genomic analysis of phage 2972 with the six other sequenced S. thermophilus phage genomes confirmed that the replication module is conserved and that cos- and pac-type phages have distinct structural and packaging genes. Two group I introns were identified in the genome of 2972. They interrupted the genes coding for the putative endolysin and the terminase large subunit. Phage mRNA splicing was demonstrated for both introns, and the secondary structures were predicted. Eight structural proteins were also identified by N-terminal sequencing and/or matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Detailed analysis of the putative minor tail proteins ORF19 and ORF21 as well as the putative receptor-binding protein ORF20 showed the following interesting features: (i) ORF19 is a hybrid protein, because it displays significant identity with both pac- and cos-type phages; (ii) ORF20 is unique; and (iii) a protein similar to ORF21 of 2972 was also found in the structure of the cos-type phage DT1, indicating that this structural protein is present in both S. thermophilus phage groups. The implications of these findings for phage classification are discussed.
Figures




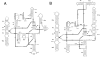

References
-
- Beck, K., and B. Brodsky. 1998. Supercoiled protein motifs: the collagen triple-helix and the α-helical coiled coil. J. Struct. Biol. 122:17-29. - PubMed
-
- Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarelle, M.-Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. M. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of a group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

