Proton sensitivity of ASIC1 appeared with the rise of fishes by changes of residues in the region that follows TM1 in the ectodomain of the channel
- PMID: 16002453
- PMCID: PMC1464184
- DOI: 10.1113/jphysiol.2005.087734
Proton sensitivity of ASIC1 appeared with the rise of fishes by changes of residues in the region that follows TM1 in the ectodomain of the channel
Abstract
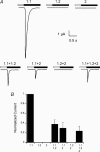
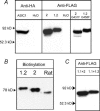
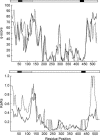
The acid-sensitive ion channel 1 (ASIC1) is a neuronal Na+ channel insensitive to changes in membrane potential but is gated by external protons. Proton sensitivity is believed to be essential for the role of ASIC1 in modulating synaptic transmission and nociception in the mammalian nervous system. To examine the structural determinants that confer proton sensitivity, we cloned and functionally characterized ASIC1 from different species of the chordate lineage: lamprey, shark, toadfish and chicken. We observed that ASIC1s from early vertebrates (lamprey and shark) were proton insensitive in spite of a high degree of amino acid conservation (66-67%) with their mammalian counterparts. Sequence analysis showed that proton-sensitive ASIC1s could not be distinguished from proton-insensitive channels by any signature in the protein sequence. Chimeras made with rat ASIC1 (rASIC1) and lamprey or shark indicated that most of the ectodomain of rASIC1 was required to confer proton sensitivity and the distinct kinetics of activation and desensitization of the rat channel. Proton-sensitive chimeras contained the segment D78-E136, together with residues D351, Q358 and E359 of the rat sequence. However, none of the functional chimeras containing only part of the rat extracellular domain retained the kinetics of activation and desensitization of rASIC1, suggesting that residues distributed in several regions of the ectodomain contribute to allosteric changes underlying activation and desensitization. The results also demonstrate that gating by protons is not a feature common to all ASIC1 channels. Proton sensitivity arose recently in evolution, implying that agonists different from protons activate ASIC1 in lower vertebrates.
Figures

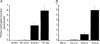



References
-
- Babini E, Paukert M, Geisler HS, Gründer S. Alternative splicing and interaction with di- and polyvalent cations control the dynamic range of acid-sensing ion channel 1 (ASIC1) J Biol Chem. 2002;277:41597–41603. - PubMed
-
- Champigny G, Voilley N, Waldmann R, Lazdunski M. Mutations causing neurodegeneration in Caenorhabditis elegans drastically alter the pH sensitivity and inactivation of the mammalian H+-gated Na+ channel MDEG1. J Biol Chem. 1998;273:15418–15422. - PubMed
-
- Coric T, Zhang P, Todorovic N, Canessa CM. The extracellular domain determines the kinetics of desensitization in acid-sensitive ion channel 1. J Biol Chem. 2003;278:45240–45247. - PubMed
-
- Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

