The genetics of regulatory variation in the human genome
- PMID: 16004727
- PMCID: PMC3525257
- DOI: 10.1186/1479-7364-2-2-126
The genetics of regulatory variation in the human genome
Abstract
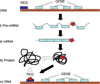
The regulation of gene expression plays an important role in complex phenotypes, including disease in humans. For some genes, the genetic mechanisms influencing gene expression are well elucidated; however, it is unclear how applicable these results are to gene expression on a genome-wide level. Studies in model organisms and humans have clearly documented gene expression variation among individuals and shown that a significant proportion of this variation has a genetic basis. Recent studies combine microarray surveys of gene expression for thousands of genes with dense marker maps, and are beginning to identify regions in the human genome that have functional effects on gene expression. This paper reviews recent developments and methodologies in this field, and discusses implications and future directions of this research in the context of understanding the influence of human genomic variation on the regulation of gene expression.
Figures
References
-
- Wray GA. Transcriptional regulation and the evolution of development. Int J Dev Biol. 2003;47:675–684. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

