Metabolic engineering of isoflavonoid biosynthesis in alfalfa
- PMID: 16006598
- PMCID: PMC1183411
- DOI: 10.1104/pp.105.062539
Metabolic engineering of isoflavonoid biosynthesis in alfalfa
Abstract
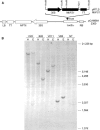
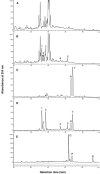
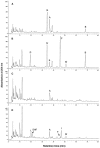
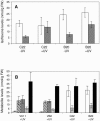
The potential health benefits of dietary isoflavones have generated considerable interest in engineering the synthesis of these phytoestrogens into plants. Genistein glucoside production (up to 50 nmol g(-1) fresh weight) was engineered in alfalfa (Medicago sativa) leaves by constitutive expression of isoflavone synthase from Medicago truncatula (MtIFS1). Glucosides of biochanin A (4'-O-methylgenistein) and pratensein (3'-hydroxybiochanin A) also accumulated. Although MtIFS1 was highly expressed in all organs examined, genistein accumulation was limited to leaves. MtIFS1-expressing lines accumulated several additional isoflavones, including formononetin and daidzein, in response to UV-B or Phoma medicaginis, whereas the chalcone and flavanone precursors of these compounds accumulated in control lines. Enhanced accumulation of the phytoalexin medicarpin was observed in P. medicaginis-infected leaves of MtIFS1-expressing plants. Microarray profiling indicated that MtIFS1 expression does not significantly alter global gene expression in the leaves. Our results highlight some of the challenges associated with metabolic engineering of plant natural products, including tissue-specific accumulation, potential for further modification by endogenous enzyme activities (hydroxylation, methylation, and glycosylation), and the differential response of engineered plants to environmental factors.
Figures




Similar articles
-
Multigene synergism increases the isoflavone and proanthocyanidin contents of Medicago truncatula.Plant Biotechnol J. 2016 Mar;14(3):915-25. doi: 10.1111/pbi.12445. Epub 2015 Aug 11. Plant Biotechnol J. 2016. PMID: 26260843 Free PMC article.
-
Changes in the profile of flavonoid accumulation in Medicago truncatula leaves during infection with fungal pathogen Phoma medicaginis.Plant Physiol Biochem. 2009 Sep;47(9):847-53. doi: 10.1016/j.plaphy.2009.05.004. Epub 2009 May 28. Plant Physiol Biochem. 2009. PMID: 19541494
-
Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures.Plant Physiol. 2008 Feb;146(2):387-402. doi: 10.1104/pp.107.108431. Epub 2007 Nov 30. Plant Physiol. 2008. PMID: 18055588 Free PMC article.
-
Plants as bioreactors: a comparative study suggests that Medicago truncatula is a promising production system.J Biotechnol. 2005 Oct 17;120(1):121-34. doi: 10.1016/j.jbiotec.2005.04.026. Epub 2005 Jul 18. J Biotechnol. 2005. PMID: 16026877 Review.
-
Genetic and metabolic engineering of isoflavonoid biosynthesis.Appl Microbiol Biotechnol. 2010 May;86(5):1293-312. doi: 10.1007/s00253-010-2512-8. Epub 2010 Mar 23. Appl Microbiol Biotechnol. 2010. PMID: 20309543 Review.
Cited by
-
Genome sequencing and genome resources in model legumes.Plant Physiol. 2007 Jun;144(2):588-93. doi: 10.1104/pp.107.097493. Plant Physiol. 2007. PMID: 17556522 Free PMC article. Review. No abstract available.
-
Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula.Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):17909-15. doi: 10.1073/pnas.0708697104. Epub 2007 Oct 30. Proc Natl Acad Sci U S A. 2007. PMID: 17971436 Free PMC article.
-
Engineered coumarin accumulation reduces mycotoxin-induced oxidative stress and disease susceptibility.Plant Biotechnol J. 2023 Dec;21(12):2490-2506. doi: 10.1111/pbi.14144. Epub 2023 Aug 14. Plant Biotechnol J. 2023. PMID: 37578146 Free PMC article.
-
Engineering isoflavone metabolism with an artificial bifunctional enzyme.Planta. 2006 Aug;224(3):496-507. doi: 10.1007/s00425-006-0233-0. Epub 2006 Feb 16. Planta. 2006. PMID: 16482434
-
The Effects of Domestication on Secondary Metabolite Composition in Legumes.Front Genet. 2020 Sep 18;11:581357. doi: 10.3389/fgene.2020.581357. eCollection 2020. Front Genet. 2020. PMID: 33193705 Free PMC article. Review.
References
-
- Akashi T, Sawada Y, Shimada H, Sakurai N, Aoki T, Ayabe S (2003) cDNA cloning and biochemical characterization of S-adenosyl-L-methionine: 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol 44: 103–112 - PubMed
-
- Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y (1987) Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem 262: 5592–5595 - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current Protocols in Molecular Biology. John Wiley and Sons, New York
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

