DNA lesions induced by UV A1 and B radiation in human cells: comparative analyses in the overall genome and in the p53 tumor suppressor gene
- PMID: 16009942
- PMCID: PMC1174921
- DOI: 10.1073/pnas.0502311102
DNA lesions induced by UV A1 and B radiation in human cells: comparative analyses in the overall genome and in the p53 tumor suppressor gene
Abstract
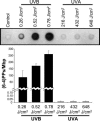
The UV components of sunlight (UVA and UVB) are implicated in the etiology of human skin cancer. The underlying mechanism of action for UVB carcinogenicity is well defined; however, the mechanistic involvement of UVA in carcinogenesis is not fully delineated. We investigated the genotoxicity of UVA1 versus UVB in the overall genome and in the p53 tumor suppressor gene in normal human skin fibroblasts. Immuno-dot blot analysis identified the cis-syn cyclobutane pyrimidine-dimer (CPD) as a distinctive UVB-induced lesion and confirmed its formation in the genomic DNA of UVA1-irradiated cells dependent on radiation dose. HPLC/tandem MS analysis showed an induction of 8-oxo-7,8-dihydro-2'-deoxyguanosine in the genomic DNA of UVA1-irradiated cells only. Mapping of DNA damages by terminal transferase-dependent PCR revealed preferential, but not identical, formation of polymerase-blocking lesions and/or strand breaks along exons 5-8 of the p53 gene in UVB- and UVA1-irradiated cells. The UVB-induced lesions detected by terminal transferase-PCR were almost exclusively mapped to pyrimidine-rich sequences; however, the UVA1-induced lesions were mapped to purine- and pyrimidine-containing sequences along the p53 gene. Cleavage assays with lesion-specific DNA repair enzymes coupled to ligation-mediated PCR showed preferential, but not identical, formation of CPDs along the p53 gene in UVB- and UVA1-irradiated cells. Additionally, dose-dependent formation of oxidized and ring-opened purines and abasic sites was established in the p53 gene in only UVA1-irradiated cells. We conclude that UVA1 induces promutagenic CPDs and oxidative DNA damage at both the genomic and nucleotide resolution level in normal human skin fibroblasts.
Figures



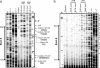
References
-
- Woodhead, A. D., Setlow, R. B. & Tanaka, M. (1999) J. Epidemiol. 9, S102-S114. - PubMed
-
- de Gruijl, F. R. (1999) Eur. J. Cancer 35, 2003-2009. - PubMed
-
- Jhappan, C., Noonan, F. P. & Merlino, G. (2003) Oncogene 22, 3099-3112. - PubMed
-
- Pfeifer, G. P., You, Y. H. & Besaratinia, A. (2005) Mutat. Res. 571, 19-31. - PubMed
-
- Cadet, J., Sage, E. & Douki, T. (2005) Mutat. Res. 571, 3-17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

