Structural determinants of alpha4beta2 nicotinic acetylcholine receptor trafficking
- PMID: 16014729
- PMCID: PMC6725434
- DOI: 10.1523/JNEUROSCI.1079-05.2005
Structural determinants of alpha4beta2 nicotinic acetylcholine receptor trafficking
Abstract
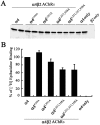
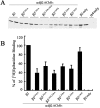
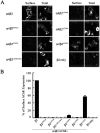
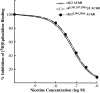
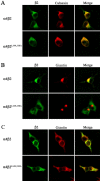
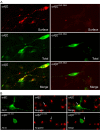
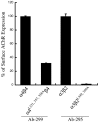
The structural determinants of nicotinic acetylcholine receptor (AChR) trafficking have yet to be fully elucidated. Hydrophobic residues occur within short motifs important for endoplasmic reticulum (ER) export or endocytotic trafficking. Hence, we tested whether highly conserved hydrophobic residues, primarily leucines, in the cytoplasmic domain of the alpha4beta2 AChR subunits were required for cell surface expression of alpha4beta2 AChRs. Mutation of F350, L351, L357, and L358 to alanine in the alpha4 AChR subunit attenuates cell surface expression of mutant alpha4beta2 AChRs. Mutation of F342, L343, L349, and L350 to alanine at homologous positions in the beta2 AChR subunit abolishes cell surface expression of mutant alpha4beta2 AChRs. The hydrophobic nature of the leucine residue is a primary determinant of its function because mutation of L343 to another hydrophobic amino acid, phenylalanine, in the beta2 AChR subunit only poorly inhibits trafficking of mutant alpha4beta2 AChR to the cell surface. All mutant alpha4beta2 AChRs exhibit high-affinity binding for [3H]epibatidine. In both tsA201 cells and differentiated SH-SY5Y neural cells, wild-type alpha4beta2 AChRs colocalize with the Golgi marker giantin, whereas mutant alpha4beta2 AChRs fail to do so. The striking difference between mutant alpha4 versus mutant beta2 AChR subunits on cell surface expression of mutant alpha4beta2 AChRs points to a cooperative or regulatory role for the alpha4 AChR subunit and an obligatory role for the beta2 AChR subunit in ER export. Collectively, our results identify, for the first time, residues within AChR subunits that are essential structural determinants of alpha4beta2 AChR ER export.
Figures


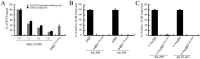
Similar articles
-
Human alpha4beta2 acetylcholine receptors formed from linked subunits.J Neurosci. 2003 Oct 8;23(27):9004-15. doi: 10.1523/JNEUROSCI.23-27-09004.2003. J Neurosci. 2003. PMID: 14534234 Free PMC article.
-
The chaperone protein 14-3-3eta interacts with the nicotinic acetylcholine receptor alpha 4 subunit. Evidence for a dynamic role in subunit stabilization.J Biol Chem. 2001 Jul 27;276(30):28281-90. doi: 10.1074/jbc.M011549200. Epub 2001 May 14. J Biol Chem. 2001. PMID: 11352901
-
Roles of accessory subunits in alpha4beta2(*) nicotinic receptors.Mol Pharmacol. 2008 Jul;74(1):132-43. doi: 10.1124/mol.108.046789. Epub 2008 Apr 1. Mol Pharmacol. 2008. PMID: 18381563
-
Localization of agonist and competitive antagonist binding sites on nicotinic acetylcholine receptors.Neurochem Int. 2000 Jun;36(7):595-645. doi: 10.1016/s0197-0186(99)00154-0. Neurochem Int. 2000. PMID: 10771117 Review.
-
Nicotinic acetylcholine receptor redux: Discovery of accessories opens therapeutic vistas.Science. 2021 Aug 13;373(6556):eabg6539. doi: 10.1126/science.abg6539. Science. 2021. PMID: 34385370 Review.
Cited by
-
Identification of sequence motifs that target neuronal nicotinic receptors to dendrites and axons.J Neurosci. 2006 Sep 20;26(38):9780-93. doi: 10.1523/JNEUROSCI.0840-06.2006. J Neurosci. 2006. PMID: 16988049 Free PMC article.
-
A highly conserved cytoplasmic cysteine residue in the α4 nicotinic acetylcholine receptor is palmitoylated and regulates protein expression.J Biol Chem. 2012 Jun 29;287(27):23119-27. doi: 10.1074/jbc.M111.328294. Epub 2012 May 16. J Biol Chem. 2012. PMID: 22593584 Free PMC article.
-
α7 nicotinic acetylcholine receptor upregulation by anti-apoptotic Bcl-2 proteins.Nat Commun. 2019 Jun 21;10(1):2746. doi: 10.1038/s41467-019-10723-x. Nat Commun. 2019. PMID: 31227712 Free PMC article.
-
Differential regulation of alpha7 nicotinic receptor gene (CHRNA7) expression in schizophrenic smokers.J Mol Neurosci. 2010 Jan;40(1-2):185-95. doi: 10.1007/s12031-009-9233-4. Epub 2009 Aug 13. J Mol Neurosci. 2010. PMID: 19680823 Free PMC article.
-
Presynaptic targeting of alpha4beta 2 nicotinic acetylcholine receptors is regulated by neurexin-1beta.J Biol Chem. 2009 Aug 28;284(35):23251-9. doi: 10.1074/jbc.M109.017384. Epub 2009 Jun 30. J Biol Chem. 2009. PMID: 19567877 Free PMC article.
References
-
- Alder NN, Johnson AE (2004) Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J Biol Chem 279: 22787-22790. - PubMed
-
- Aridor M, Traub LM (2002) Cargo selection in vesicular transport: the making and breaking of a coat. Traffic 3: 537-546. - PubMed
-
- Barlowe C (2003) Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol 13: 295-300. - PubMed
-
- Blount P, Merlie JP (1988) Native folding of an acetylcholine receptor alpha subunit expressed in the absence of other receptor subunits. J Biol Chem 263: 1072-1080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
