Humoral immune response recognizes a complex set of epitopes on human papillomavirus type 6 l1 capsomers
- PMID: 16014913
- PMCID: PMC1181614
- DOI: 10.1128/JVI.79.15.9503-9514.2005
Humoral immune response recognizes a complex set of epitopes on human papillomavirus type 6 l1 capsomers
Abstract
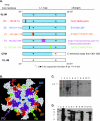
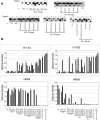
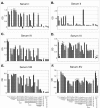
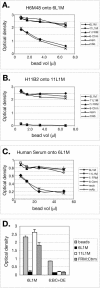
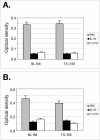
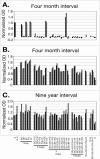
Although epitope mapping has identified residues on the human papillomavirus (HPV) major capsid protein (L1) that are important for binding mouse monoclonal antibodies, epitopes recognized by human antibodies are not known. To map epitopes on HPV type 6 (HPV6) L1, surface-exposed loops were mutated to the corresponding sequence of HPV11 L1. HPV6 L1 capsomers had one to six regions mutated, including the BC, DE, EF, FG, and HI loops and the 139 C-terminal residues. After verifying proper conformation, hybrid capsomers were used in enzyme-linked immunosorbent assays with 36 HPV6-seropositive sera from women enrolled in a study of incident HPV infection. Twelve sera were HPV6 specific, while the remainder reacted with both HPV6 and HPV11 L1. By preadsorption studies, 6/11 of these sera were shown to be cross-reactive. Among the HPV6-specific sera there was no immunodominant epitope recognized by all sera. Six of the 12 sera recognized epitopes that contained residues from combinations of the BC, DE, and FG loops, one serum recognized an epitope that consisted partially of the C-terminal arm, and three sera recognized complex epitopes to which reactivity was eliminated by switching all five loops. Reactivity in two sera was not eliminated even with all six regions swapped. The patterns of epitope recognition did not change over time in women whose sera were examined 9 years after their first-seropositive visit.
Figures

References
-
- Breitburd, F., R. Kirnbauer, N. L. Hubbert, B. Nonnenmacher, C. Trin-Dinh-Desmarquet, G. Orth, J. T. Schiller, and D. R. Lowy. 1995. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J. Virol. 69:3959-3963. - PMC - PubMed
-
- Caparros-Wanderley, W., N. Savage, M. Hill-Perkins, G. Layton, J. Weber, and D. H. Davies. 1999. Intratype sequence variation among clinical isolates of the human papillomavirus type 6 L1 ORF: clustering of mutations and identification of a frequent amino acid sequence variant. J. Gen. Virol. 80:1025-1033. - PubMed
-
- Carter, J. J., L. A. Koutsky, J. P. Hughes, S. K. Lee, J. Kuypers, N. Kiviat, and D. A. Galloway. 2000. Comparison of human papillomavirus types 16, 18, and 6 capsid antibody responses following incident infection. J. Infect. Dis. 181:1911-1919. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

