The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in gag that regulate nuclear localization and particle formation
- PMID: 16014916
- PMCID: PMC1181613
- DOI: 10.1128/JVI.79.15.9540-9555.2005
The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in gag that regulate nuclear localization and particle formation
Abstract
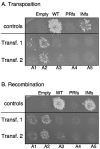
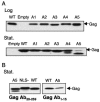
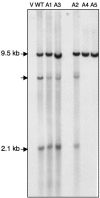
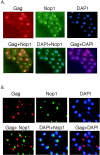
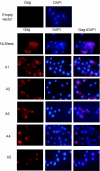
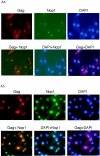
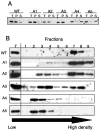
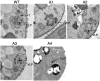
Tf1 is a long terminal repeat-containing retrotransposon of Schizosaccharomyces pombe that is studied to further our understanding of retrovirus propagation. One important application is to examine Tf1 as a model for how human immunodeficiency virus type 1 proteins enter the nucleus. The accumulation of Tf1 Gag in the nucleus requires an N-terminal nuclear localization signal (NLS) and the nuclear pore factor Nup124p. Here, we report that NLS activity is regulated by adjacent residues. Five mutant transposons were made, each with sequential tracts of four amino acids in Gag replaced by alanines. All five versions of Tf1 transposed with frequencies that were significantly lower than that of the wild type. Although all five made normal amounts of Gag, two of the mutations did not make cDNA, indicating that Gag contributed to reverse transcription. The localization of the Gag in the nucleus was significantly reduced by mutations A1, A2, and A3. These results identified residues in Gag that contribute to the function of the NLS. The Gags of A4 and A5 localized within the nucleus but exhibited severe defects in the formation of virus-like particles. Of particular interest was that the mutations in Gag-A4 and Gag-A5 caused their nuclear localization to become independent of Nup124p. These results suggested that Nup124p was only required for import of Tf1 Gag because of its extensive multimerization.
Figures



Similar articles
-
Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p.Mol Cell Biol. 2000 Oct;20(20):7798-812. doi: 10.1128/MCB.20.20.7798-7812.2000. Mol Cell Biol. 2000. PMID: 11003674 Free PMC article.
-
The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr.Mol Biol Cell. 2005 Apr;16(4):1823-38. doi: 10.1091/mbc.e04-07-0583. Epub 2005 Jan 19. Mol Biol Cell. 2005. PMID: 15659641 Free PMC article.
-
Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1.Mol Cell Biol. 1999 Aug;19(8):5768-84. doi: 10.1128/MCB.19.8.5768. Mol Cell Biol. 1999. PMID: 10409764 Free PMC article.
-
Nuclear localization signals and human disease.IUBMB Life. 2009 Jul;61(7):697-706. doi: 10.1002/iub.194. IUBMB Life. 2009. PMID: 19514019 Review.
-
Nucleocytoplasmic shuttling of phospholipase C-delta1: a link to Ca2+.J Cell Biochem. 2006 Feb 1;97(2):233-43. doi: 10.1002/jcb.20677. J Cell Biochem. 2006. PMID: 16240320 Review.
Cited by
-
Additional ORFs in Plant LTR-Retrotransposons.Front Plant Sci. 2020 May 26;11:555. doi: 10.3389/fpls.2020.00555. eCollection 2020. Front Plant Sci. 2020. PMID: 32528484 Free PMC article. Review.
-
The chromodomain of Tf1 integrase promotes binding to cDNA and mediates target site selection.J Virol. 2009 Mar;83(6):2675-85. doi: 10.1128/JVI.01588-08. Epub 2008 Dec 24. J Virol. 2009. PMID: 19109383 Free PMC article.
-
The Long Terminal Repeat Retrotransposons Tf1 and Tf2 of Schizosaccharomyces pombe.Microbiol Spectr. 2015 Aug;3(4):10.1128/microbiolspec.MDNA3-0040-2014. doi: 10.1128/microbiolspec.MDNA3-0040-2014. Microbiol Spectr. 2015. PMID: 26350316 Free PMC article. Review.
-
New insights into the nuclear localization of retroviral Gag proteins.Nucleus. 2011 Mar-Apr;2(2):92-7. doi: 10.4161/nucl.2.2.15018. Nucleus. 2011. PMID: 21738831 Free PMC article.
-
Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity.PLoS Pathog. 2013;9(10):e1003693. doi: 10.1371/journal.ppat.1003693. Epub 2013 Oct 10. PLoS Pathog. 2013. PMID: 24130490 Free PMC article.
References
-
- Allen, N. P. C., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268-29274. - PubMed
-
- Allen, N. P. C., S. S. Patel, L. Huang, R. F. Chalkley, A. Burlingame, M. Lutzmann, E. C. Hurt, and M. Rexach. 2002. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1:930-946. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

