Suppression of RNA interference by adenovirus virus-associated RNA
- PMID: 16014917
- PMCID: PMC1181602
- DOI: 10.1128/JVI.79.15.9556-9565.2005
Suppression of RNA interference by adenovirus virus-associated RNA
Abstract
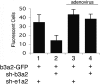
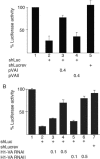
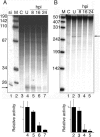
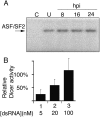
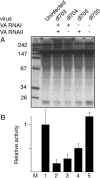
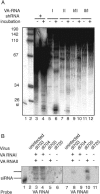
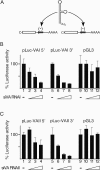
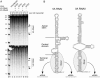
We show that human adenovirus inhibits RNA interference (RNAi) at late times of infection by suppressing the activity of two key enzyme systems involved, Dicer and RNA-induced silencing complex (RISC). To define the mechanisms by which adenovirus blocks RNAi, we used a panel of mutant adenoviruses defective in virus-associated (VA) RNA expression. The results show that the virus-associated RNAs, VA RNAI and VA RNAII, function as suppressors of RNAi by interfering with the activity of Dicer. The VA RNAs bind Dicer and function as competitive substrates squelching Dicer. Further, we show that VA RNAI and VA RNAII are processed by Dicer, both in vitro and during a lytic infection, and that the resulting short interfering RNAs (siRNAs) are incorporated into active RISC. Dicer cleaves the terminal stem of both VA RNAI and VA RNAII. However, whereas both strands of the VA RNAI-specific siRNA are incorporated into RISC, the 3' strand of the VA RNAII-specific siRNA is selectively incorporated during a lytic infection. In summary, our work shows that adenovirus suppresses RNAi during a lytic infection and gives insight into the mechanisms of RNAi suppression by VA RNA.
Figures
References
-
- Akusjärvi, G., and J. Stevenin. 2003. Remodelling of the host cell RNA splicing machinery during an adenovirus infection. Curr. Top. Microbiol. Immunol. 272:253-286. - PubMed
-
- Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

