mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A
- PMID: 16014927
- PMCID: PMC1181552
- DOI: 10.1128/JVI.79.15.9651-9664.2005
mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A
Abstract
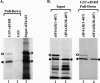
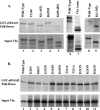
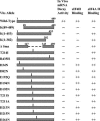
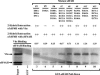
During lytic infections, the virion host shutoff (Vhs) protein of herpes simplex virus accelerates the degradation of both host and viral mRNAs. In so doing, it helps redirect the cell from host to viral protein synthesis and facilitates the sequential expression of different viral genes. Vhs interacts with the cellular translation initiation factor eIF4H, and several point mutations that abolish its mRNA degradative activity also abrogate its ability to bind eIF4H. In addition, a complex containing bacterially expressed Vhs and a glutathione S-transferase (GST)-eIF4H fusion protein has RNase activity. eIF4H shares a region of sequence homology with eIF4B, and it appears to be functionally similar in that both stimulate the RNA helicase activity of eIF4A, a component of the mRNA cap-binding complex eIF4F. We show that eIF4H interacts physically with eIF4A in the yeast two-hybrid system and in GST pull-down assays and that the two proteins can be coimmunoprecipitated from mammalian cells. Vhs also interacts with eIF4A in GST pull-down and coimmunoprecipitation assays. Site-directed mutagenesis of Vhs and eIF4H revealed residues of each that are important for their mutual interaction, but not for their interaction with eIF4A. Thus, Vhs, eIF4H, and eIF4A comprise a group of proteins, each of which is able to interact directly with the other two. Whether they interact simultaneously as a tripartite complex or sequentially is unclear. The data suggest a mechanism for linking the degradation of an mRNA to its translation and for targeting Vhs to mRNAs and to regions of translation initiation.
Figures





Similar articles
-
mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease.J Virol. 2002 Sep;76(17):8560-71. doi: 10.1128/jvi.76.17.8560-8571.2002. J Virol. 2002. PMID: 12163576 Free PMC article.
-
Small interfering RNAs that deplete the cellular translation factor eIF4H impede mRNA degradation by the virion host shutoff protein of herpes simplex virus.J Virol. 2008 Jul;82(13):6600-9. doi: 10.1128/JVI.00137-08. Epub 2008 Apr 30. J Virol. 2008. PMID: 18448541 Free PMC article.
-
mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor.J Virol. 2001 Nov;75(21):10272-80. doi: 10.1128/JVI.75.21.10272-10280.2001. J Virol. 2001. PMID: 11581395 Free PMC article.
-
Early shutoff of host protein synthesis in cells infected with herpes simplex viruses.Acta Virol. 2001;45(5-6):269-77. Acta Virol. 2001. PMID: 12083325 Review.
-
Virus-encoded endonucleases: expected and novel functions.Wiley Interdiscip Rev RNA. 2013 Nov-Dec;4(6):693-708. doi: 10.1002/wrna.1188. Epub 2013 Jul 30. Wiley Interdiscip Rev RNA. 2013. PMID: 23900973 Review.
Cited by
-
The herpes simplex virus 1 virion host shutoff protein enhances translation of viral late mRNAs by preventing mRNA overload.J Virol. 2014 Sep 1;88(17):9624-32. doi: 10.1128/JVI.01350-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920814 Free PMC article.
-
Roles of nucleic acid substrates and cofactors in the vhs protein activity of pseudorabies virus.Vet Res. 2015 Dec 24;46:141. doi: 10.1186/s13567-015-0284-y. Vet Res. 2015. PMID: 26704628 Free PMC article.
-
'Ribozoomin'--translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs).Curr Protein Pept Sci. 2012 Jun;13(4):305-30. doi: 10.2174/138920312801619385. Curr Protein Pept Sci. 2012. PMID: 22708493 Free PMC article. Review.
-
Multiple Posttranscriptional Strategies To Regulate the Herpes Simplex Virus 1 vhs Endoribonuclease.J Virol. 2018 Aug 16;92(17):e00818-18. doi: 10.1128/JVI.00818-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29925667 Free PMC article.
-
Nuclear-cytoplasmic compartmentalization of the herpes simplex virus 1 infected cell transcriptome is co-ordinated by the viral endoribonuclease vhs and cofactors to facilitate the translation of late proteins.PLoS Pathog. 2018 Nov 26;14(11):e1007331. doi: 10.1371/journal.ppat.1007331. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30475899 Free PMC article.
References
-
- Becker, Y., E. Tavor, Y. Asher, C. Berkowiltz, and M. Moyal. 1993. Effect of herpes simplex virus type-1 UL41 gene on the stability of mRNA from the cellular genes: beta-actin, fibronectin, glucose transporter-1, and docking protein, and on virus intraperitoneal pathogenicity of newborn mice. Virus Genes 7:133-143. - PubMed
-
- Callaghan, A. J., J. P. Aurikko, L. L. Ilag, G. J. Gunter, V. Chandran, K. Kuhnel, L. Poljak, A. J. Carpousis, C. V. Robinson, M. F. Symmons, and B. F. Luisi. 2004. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 340:965-979. - PubMed
-
- Carpousis, A. J. 2002. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 30:150-155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

