The capsid protein of satellite Panicum mosaic virus contributes to systemic invasion and interacts with its helper virus
- PMID: 16014937
- PMCID: PMC1181559
- DOI: 10.1128/JVI.79.15.9756-9764.2005
The capsid protein of satellite Panicum mosaic virus contributes to systemic invasion and interacts with its helper virus
Abstract
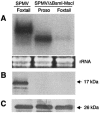
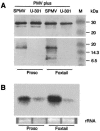
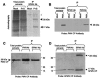
Satellite panicum mosaic virus (SPMV) depends on its helper Panicum mosaic virus (PMV) for replication and spread in host plants. The SPMV RNA encodes a 17-kDa capsid protein (CP) that is essential for formation of its 16-nm virions. The results of this study indicate that in addition to the expression of the full-length SPMV CP from the 5'-proximal AUG start codon, SPMV RNA also expresses a 9.4-kDa C-terminal protein from the third in-frame start codon. Differences in solubility between the full-length protein and its C-terminal product were observed. Subcellular fractionation of infected plant tissues showed that SPMV CP accumulates in the cytosol, cell wall-, and membrane-enriched fractions. However, the 9.4-kDa protein exclusively cofractionated with cell wall- and membrane-enriched fractions. Earlier studies revealed that the 5'-untranslated region (5'-UTR) from nucleotides 63 to 104 was associated with systemic infection in a host-specific manner in millet plants. This study shows that nucleotide deletions and insertions in the 5'-UTR plus simultaneous truncation of the N-terminal part of the CP impaired SPMV spread in foxtail millet, but not in proso millet plants. In contrast, the expression of the full-length version of SPMV CP efficiently compensated the negative effect of the 5'-UTR deletions in foxtail millet. Finally, immunoprecipitation assays revealed the presence of a specific interaction between the capsid proteins of SPMV and its helper virus (PMV). Our findings show that the SPMV CP has several biological functions, including facilitating efficient satellite virus infection and movement in millet plants.
Figures



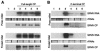
References
-
- Batten, J. S. 2002. Replication and translation of panicum mosaic virus. Ph.D. dissertation. Texas A&M University, College Station.
-
- Buzen, F. G., C. L. Niblett, G. R. Hooper, J. Hubbard, and M. A. Newman. 1984. Further characterization of panicum mosaic virus and its associated satellite virus. Phytopathology 74:313-318.
-
- Callaway, A., D. Giesman-Cookmeyer, E. T. Gillock, T. L. Sit, and S. A. Lommel. 2001. The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39:419-460. - PubMed
-
- Chapman, S., T. Kavanagh, and D. Baulcombe. 1992. Potato virus X as a vector for gene expression in plants. Plant J. 2:549-557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

