Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition
- PMID: 16014940
- PMCID: PMC1181564
- DOI: 10.1128/JVI.79.15.9786-9798.2005
Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition
Abstract
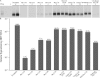
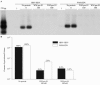
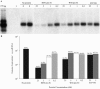
Hepatitis B virus (HBV) and woolly monkey hepatitis B virus (WMHBV) are primate hepadnaviruses that display restricted tissue and host tropisms. Hepatitis D virus (HDV) particles pseudotyped with HBV and WMHBV envelopes (HBV-HDV and WM-HDV) preferentially infect human and spider monkey hepatocytes, respectively, thereby confirming host range bias in vitro. The analysis of chimeric HBV and WMHBV large (L) envelope proteins suggests that the pre-S1 domain may comprise two regions that affect infectivity: one within the amino-terminal 40 amino acids of pre-S1 and one downstream of this region. In the present study, we further characterized the role of the amino terminus of pre-S1 in infectivity by examining the ability of synthetic peptides to competitively block HDV infection of primary human and spider monkey hepatocytes. A synthetic peptide representing the first 45 residues of the pre-S1 domain of the HBV L protein blocked infectivity of HBV-HDV and WM-HDV, with a requirement for myristylation of the amino terminal residue. Competition studies with truncated peptides suggested that pre-S1 residues 5 to 20 represent the minimal domain for inhibition of HDV infection and, thus, presumably represent the residues involved in virus-host receptor interaction. Recombinant pre-S1 proteins expressed in insect cells blocked infection with HBV-HDV and WM-HDV at a concentration of 1 nanomolar. The ability of short pre-S1 peptides to efficiently inhibit HDV infection suggests that they represent suitable ligands for identification of the HBV receptor and that a pre-S1 mimetic may represent a rational therapy for the treatment of HBV infection.
Figures
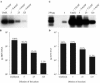
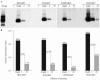
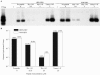


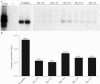
References
-
- Barrera, A., and R. E. Lanford. 2004. Infection of primary chimpanzee hepatocytes with recombinant hepatitis D virus particles: a surrogate model for hepatitis B virus. Methods Mol. Med. 95:131-142. - PubMed
-
- Bruss, V., J. Hagelsten, E. Gerhardt, and P. R. Galle. 1996. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218:396-399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

