A peptide derived from RNA recognition motif 2 of human la protein binds to hepatitis C virus internal ribosome entry site, prevents ribosomal assembly, and inhibits internal initiation of translation
- PMID: 16014945
- PMCID: PMC1181605
- DOI: 10.1128/JVI.79.15.9842-9853.2005
A peptide derived from RNA recognition motif 2 of human la protein binds to hepatitis C virus internal ribosome entry site, prevents ribosomal assembly, and inhibits internal initiation of translation
Abstract
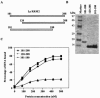
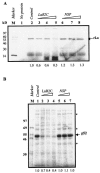
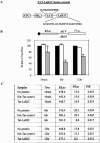
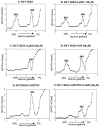
Human La protein is known to interact with hepatitis C virus (HCV) internal ribosome entry site (IRES) and stimulate translation. Previously, we demonstrated that mutations within HCV SL IV lead to reduced binding to La-RNA recognition motif 2 (RRM2) and drastically affect HCV IRES-mediated translation. Also, the binding of La protein to SL IV of HCV IRES was shown to impart conformational alterations within the RNA so as to facilitate the formation of functional initiation complex. Here, we report that a synthetic peptide, LaR2C, derived from the C terminus of La-RRM2 competes with the binding of cellular La protein to the HCV IRES and acts as a dominant negative inhibitor of internal initiation of translation of HCV RNA. The peptide binds to the HCV IRES and inhibits the functional initiation complex formation. An Huh7 cell line constitutively expressing a bicistronic RNA in which both cap-dependent and HCV IRES-mediated translation can be easily assayed has been developed. The addition of purified TAT-LaR2C recombinant polypeptide that allows direct delivery of the peptide into the cells showed reduced expression of HCV IRES activity in this cell line. The study reveals valuable insights into the role of La protein in ribosome assembly at the HCV IRES and also provides the basis for targeting ribosome-HCV IRES interaction to design potent antiviral therapy.
Figures



References
-
- Ali, N., G. J. M. Pruijn, D. J. Kenan, J. D. Keene, and A. Siddiqui. 2000. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 275:27531-27540. - PubMed
-
- Bartenschlager, R. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral. Hepat. 6:165-181. - PubMed
-
- Becker-Hapak, M., S. S. McAllister, and S. F. Dowdy. 2001. TAT-mediated protein transduction into mammalian cells. Methods 24:247-256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

