Geminivirus C3 protein: replication enhancement and protein interactions
- PMID: 16014949
- PMCID: PMC1181577
- DOI: 10.1128/JVI.79.15.9885-9895.2005
Geminivirus C3 protein: replication enhancement and protein interactions
Abstract
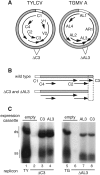
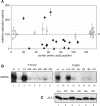
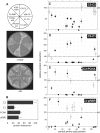
Most dicot-infecting geminiviruses encode a replication enhancer protein (C3, AL3, or REn) that is required for optimal replication of their small, single-stranded DNA genomes. C3 interacts with C1, the essential viral replication protein that initiates rolling circle replication. C3 also homo-oligomerizes and interacts with at least two host-encoded proteins, proliferating cell nuclear antigen (PCNA) and the retinoblastoma-related protein (pRBR). It has been proposed that protein interactions contribute to C3 function. Using the C3 protein of Tomato yellow leaf curl virus, we examined the impact of mutations to amino acids that are conserved across the C3 protein family on replication enhancement and protein interactions. Surprisingly, many of the mutations did not affect replication enhancement activity of C3 in tobacco protoplasts. Other mutations either enhanced or were detrimental to C3 replication activity. Analysis of mutated proteins in yeast two-hybrid assays indicated that mutations that inactivate C3 replication enhancement activity also reduce or inactivate C3 oligomerization and interaction with C1 and PCNA. In contrast, mutated C3 proteins impaired for pRBR binding are fully functional in replication assays. Hydrophobic residues in the middle of the C3 protein were implicated in C3 interaction with itself, C1, and PCNA, while polar resides at both the N and C termini of the protein are important for C3-pRBR interaction. These experiments established the importance of C3-C3, C3-C1, and C3-PCNA interactions in geminivirus replication. While C3-pRBR interaction is not required for viral replication in cycling cells, it may play a role during infection of differentiated cells in intact plants.
Figures


References
-
- Castillo, A. G., D. Collinet, S. Deret, A. Kashoggi, and E. R. Bejarano. 2003. Dual interaction of plant PCNA with geminivirus replication accessory protein (REn) and viral replication protein (Rep). Virology 312:381-394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

