Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps
- PMID: 16014954
- PMCID: PMC1181570
- DOI: 10.1128/JVI.79.15.9933-9944.2005
Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps
Abstract
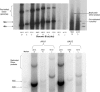
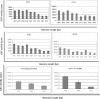
The limited packaging capacity of adeno-associated virus (AAV) precludes the design of vectors for the treatment of diseases associated with larger genes. Autonomous parvoviruses, such as minute virus of mice and B19, while identical in size (25 nm), are known to package larger genomes of 5.1 and 5.6 kb, respectively, compared to AAV genomes of 4.7 kb. One primary difference is the fact that wild-type (wt) AAV utilizes three capsid subunits instead of two to form the virion shell. In this study, we have characterized the packaging capacity of AAV serotypes 1 through 5 with and without the Vp2 subunit. Using reporter transgene cassettes that range in size from 4.4 to 6.0 kb, we determined that serotypes 1 through 5 with and without Vp2 could successfully package, replicate in, and transduce cells. Dot blot analysis established that packaging efficiency was similar for all vector cassettes and that the integrity of encapsidated genomes was intact regardless of size. Although physical characterization determined that virion structures were indistinguishable from wt, transduction experiments determined that all serotype vectors carrying larger genomes (5.3 kb and higher) transduced cells less efficiently (within a log) than AAV encapsidating wt size genomes. This result was not unique to reporter genes and was observed for CFTR vector cassettes ranging in size from 5.1 to 5.9 kb. No apparent advantage in packaging efficiency was observed when Vp2 was present or absent from the virion. Further analysis determined that a postentry step was responsible for the block in infection and specific treatment of cells upon infection with proteasome inhibitors increased transduction of AAV encapsidating larger DNA templates to wt levels, suggesting a preferential degradation of virions encapsidating larger-than-wt genomes. This study illustrates that AAV is capable of packaging and protecting recombinant genomes as large as 6.0 kb but the larger genome-containing virions are preferentially degraded by the proteasome and that this block can be overcome by the addition of proteasome inhibitors.
Figures

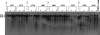

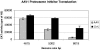

Similar articles
-
Quantitative analysis of the packaging capacity of recombinant adeno-associated virus.Hum Gene Ther. 1996 Nov 10;7(17):2101-12. doi: 10.1089/hum.1996.7.17-2101. Hum Gene Ther. 1996. PMID: 8934224
-
Improved Genome Packaging Efficiency of Adeno-associated Virus Vectors Using Rep Hybrids.J Virol. 2021 Sep 9;95(19):e0077321. doi: 10.1128/JVI.00773-21. Epub 2021 Jul 21. J Virol. 2021. PMID: 34287038 Free PMC article.
-
Self-complementary recombinant adeno-associated viral vectors: packaging capacity and the role of rep proteins in vector purity.Hum Gene Ther. 2007 Feb;18(2):171-82. doi: 10.1089/hum.2006.088. Hum Gene Ther. 2007. PMID: 17328683
-
Expressing Transgenes That Exceed the Packaging Capacity of Adeno-Associated Virus Capsids.Hum Gene Ther Methods. 2016 Feb;27(1):1-12. doi: 10.1089/hgtb.2015.140. Hum Gene Ther Methods. 2016. PMID: 26757051 Free PMC article. Review.
-
Adeno-associated virus-based vectors in gene therapy.J Biomed Sci. 2000 Jul-Aug;7(4):279-91. doi: 10.1007/BF02253246. J Biomed Sci. 2000. PMID: 10895050 Review.
Cited by
-
Advances in approaches to study cell-type specific cortical circuits throughout development.Front Cell Neurosci. 2022 Oct 17;16:1031389. doi: 10.3389/fncel.2022.1031389. eCollection 2022. Front Cell Neurosci. 2022. PMID: 36324861 Free PMC article. Review.
-
Modular dispensability of dysferlin C2 domains reveals rational design for mini-dysferlin molecules.J Biol Chem. 2012 Aug 10;287(33):27629-36. doi: 10.1074/jbc.M112.391722. Epub 2012 Jun 26. J Biol Chem. 2012. Retraction in: J Biol Chem. 2017 Jul 28;292(30):12543. doi: 10.1074/jbc.A112.391722. PMID: 22736764 Free PMC article. Retracted.
-
High-level protein production in erythroid cells derived from in vivo transduced hematopoietic stem cells.Blood Adv. 2019 Oct 8;3(19):2883-2894. doi: 10.1182/bloodadvances.2019000706. Blood Adv. 2019. PMID: 31585952 Free PMC article.
-
Emerging Issues in AAV-Mediated In Vivo Gene Therapy.Mol Ther Methods Clin Dev. 2017 Dec 1;8:87-104. doi: 10.1016/j.omtm.2017.11.007. eCollection 2018 Mar 16. Mol Ther Methods Clin Dev. 2017. PMID: 29326962 Free PMC article. Review.
-
Metabolic and Redox Signaling of the Nucleoredoxin-Like-1 Gene for the Treatment of Genetic Retinal Diseases.Int J Mol Sci. 2020 Feb 27;21(5):1625. doi: 10.3390/ijms21051625. Int J Mol Sci. 2020. PMID: 32120883 Free PMC article. Review.
References
-
- Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. - PubMed
-
- Brandenburger, A., E. Coessens, K. El Bakkouri, and T. Velu. 1999. Influence of sequence and size of DNA on packaging efficiency of parvovirus MVM-based vectors. Hum. Gene Ther. 10:1229-1238. - PubMed
-
- Carreira, A., M. Menendez, J. Reguera, J. M. Almendral, and M. G. Mateu. 2004. In vitro disassembly of a parvovirus capsid and effect on capsid stability of heterologous peptide insertions in surface loops. J. Biol. Chem. 279:6517-6525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

