Genetic analysis of the polyomavirus DnaJ domain
- PMID: 16014958
- PMCID: PMC1181550
- DOI: 10.1128/JVI.79.15.9982-9990.2005
Genetic analysis of the polyomavirus DnaJ domain
Abstract
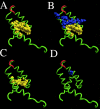
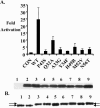
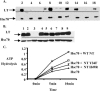
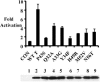
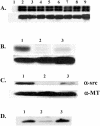
Polyomavirus T antigens share a common N-terminal sequence that comprises a DnaJ domain. DnaJ domains activate DnaK molecular chaperones. The functions of J domains have primarily been tested by mutation of their conserved HPD residues. Here, we report detailed mutagenesis of the polyomavirus J domain in both large T (63 mutants) and middle T (51 mutants) backgrounds. As expected, some J mutants were defective in binding DnaK (Hsc70); other mutants retained the ability to bind Hsc70 but were defective in stimulating its ATPase activity. Moreover, the J domain behaves differently in large T and middle T. A given mutation was twice as likely to render large T unstable as it was to affect middle T stability. This apparently arose from middle T's ability to bind stabilizing proteins such as protein phosphatase 2A (PP2A), since introduction of a second mutation preventing PP2A binding rendered some middle T J-domain mutants unstable. In large T, the HPD residues are critical for Rb-dependent effects on the host cell. Residues Q32, A33, Y34, H49, M52, and N56 within helix 2 and helix 3 of the large T J domain were also found to be required for Rb-dependent transactivation. Cyclin A promoter assays showed that J domain function also contributes to large T transactivation that is independent of Rb. Single point mutations in middle T were generally without effect. However, residue Q37 is critical for middle T's ability to form active signaling complexes. The Q37A middle T mutant was defective in association with pp60(c-src) and in transformation.
Figures
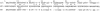


Similar articles
-
Hsc70 contacts helix III of the J domain from polyomavirus T antigens: addressing a dilemma in the chaperone hypothesis of how they release E2F from pRb.Biochemistry. 2006 Jun 6;45(22):6917-29. doi: 10.1021/bi060411d. Biochemistry. 2006. PMID: 16734427
-
Effect on polyomavirus T-antigen function of mutations in a conserved leucine-rich segment of the DnaJ domain.J Virol. 2001 Mar;75(5):2253-61. doi: 10.1128/JVI.75.5.2253-2261.2001. J Virol. 2001. PMID: 11160729 Free PMC article.
-
The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain.Mol Cell Biol. 1997 Aug;17(8):4761-73. doi: 10.1128/MCB.17.8.4761. Mol Cell Biol. 1997. PMID: 9234732 Free PMC article.
-
The role of the J domain of SV40 large T in cellular transformation.Biologicals. 1999 Mar;27(1):23-8. doi: 10.1006/biol.1998.0173. Biologicals. 1999. PMID: 10441399 Review.
-
Mutants of polyomavirus middle-T antigen.Biochim Biophys Acta. 1987 Nov 25;907(3):299-321. doi: 10.1016/0304-419x(87)90011-4. Biochim Biophys Acta. 1987. PMID: 3315000 Review.
Cited by
-
Virion assembly factories in the nucleus of polyomavirus-infected cells.PLoS Pathog. 2012;8(4):e1002630. doi: 10.1371/journal.ppat.1002630. Epub 2012 Apr 5. PLoS Pathog. 2012. PMID: 22496654 Free PMC article.
-
Structure and mechanistic insights into novel iron-mediated moonlighting functions of human J-protein cochaperone, Dph4.J Biol Chem. 2012 Apr 13;287(16):13194-205. doi: 10.1074/jbc.M112.339655. Epub 2012 Feb 24. J Biol Chem. 2012. PMID: 22367199 Free PMC article.
-
Steered molecular dynamics simulation of the binding of the bovine auxilin J domain to the Hsc70 nucleotide-binding domain.J Mol Model. 2017 Oct 23;23(11):320. doi: 10.1007/s00894-017-3453-2. J Mol Model. 2017. PMID: 29063205
-
Lessons in signaling and tumorigenesis from polyomavirus middle T antigen.Microbiol Mol Biol Rev. 2009 Sep;73(3):542-63, Table of Contents. doi: 10.1128/MMBR.00009-09. Microbiol Mol Biol Rev. 2009. PMID: 19721090 Free PMC article. Review.
-
Human Polyomaviruses: The Battle of Large and Small Tumor Antigens.Virology (Auckl). 2017 Dec 5;8:1178122X17744785. doi: 10.1177/1178122X17744785. eCollection 2017. Virology (Auckl). 2017. PMID: 29238174 Free PMC article. Review.
References
-
- Berjanskii, M. V., M. I. Riley, A. Xie, V. Semenchenko, W. R. Folk, and S. R. Van Doren. 2000. NMR structure of the N-terminal J. domain of murine polyomavirus T antigens: implications for DnaJ-like domains and for mutations of T antigens. J. Biol. Chem. 275:36094-36103. - PubMed
-
- Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. Zalvide, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

